CotH3 mediates fungal invasion of host cells during mucormycosis
- PMID: 24355926
- PMCID: PMC3871245
- DOI: 10.1172/JCI71349
CotH3 mediates fungal invasion of host cells during mucormycosis
Abstract
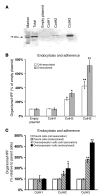
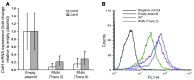
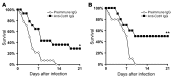
Angioinvasion is a hallmark of mucormycosis. Previously, we identified endothelial cell glucose-regulated protein 78 (GRP78) as a receptor for Mucorales that mediates host cell invasion. Here we determined that spore coat protein homologs (CotH) of Mucorales act as fungal ligands for GRP78. CotH proteins were widely present in Mucorales and absent from noninvasive pathogens. Heterologous expression of CotH3 and CotH2 in Saccharomyces cerevisiae conferred the ability to invade host cells via binding to GRP78. Homology modeling and computational docking studies indicated structurally compatible interactions between GRP78 and both CotH3 and CotH2. A mutant of Rhizopus oryzae, the most common cause of mucormycosis, with reduced CotH expression was impaired for invading and damaging endothelial cells and CHO cells overexpressing GRP78. This strain also exhibited reduced virulence in a diabetic ketoacidotic (DKA) mouse model of mucormycosis. Treatment with anti-CotH Abs abolished the ability of R. oryzae to invade host cells and protected DKA mice from mucormycosis. The presence of CotH in Mucorales explained the specific susceptibility of DKA patients, who have increased GRP78 levels, to mucormycosis. Together, these data indicate that CotH3 and CotH2 function as invasins that interact with host cell GRP78 to mediate pathogenic host-cell interactions and identify CotH as a promising therapeutic target for mucormycosis.
Figures






Comment in
-
Hostile takeover: fungal protein promotes host cell invasion.J Clin Invest. 2014 Jan;124(1):74-6. doi: 10.1172/JCI73585. Epub 2013 Dec 20. J Clin Invest. 2014. PMID: 24355916 Free PMC article.
Similar articles
-
Anti-CotH3 antibodies protect mice from mucormycosis by prevention of invasion and augmenting opsonophagocytosis.Sci Adv. 2019 Jun 12;5(6):eaaw1327. doi: 10.1126/sciadv.aaw1327. eCollection 2019 Jun. Sci Adv. 2019. PMID: 31206021 Free PMC article.
-
GRP78 and Integrins Play Different Roles in Host Cell Invasion during Mucormycosis.mBio. 2020 Jun 2;11(3):e01087-20. doi: 10.1128/mBio.01087-20. mBio. 2020. PMID: 32487760 Free PMC article.
-
The expression of fungal CotH, human glucose-regulated protein 78 (GRP78), and predicted miRNAs in macrophages and diabetic mice infected with Rhizopus oryzae.Microbiol Spectr. 2025 Jul;13(7):e0285224. doi: 10.1128/spectrum.02852-24. Epub 2025 Jun 9. Microbiol Spectr. 2025. PMID: 40488471 Free PMC article.
-
Pathogenesis of mucormycosis.Clin Infect Dis. 2012 Feb;54 Suppl 1(Suppl 1):S16-22. doi: 10.1093/cid/cir865. Clin Infect Dis. 2012. PMID: 22247441 Free PMC article. Review.
-
Host-iron assimilation: pathogenesis and novel therapies of mucormycosis.Mycoses. 2014 Dec;57 Suppl 3(0 3):13-7. doi: 10.1111/myc.12232. Epub 2014 Sep 1. Mycoses. 2014. PMID: 25178879 Free PMC article. Review.
Cited by
-
Evaluation of Sex Differences in Murine Diabetic Ketoacidosis and Neutropenic Models of Invasive Mucormycosis.J Fungi (Basel). 2021 Apr 18;7(4):313. doi: 10.3390/jof7040313. J Fungi (Basel). 2021. PMID: 33919611 Free PMC article.
-
Monoclonal antibodies: From magic bullet to precision weapon.Mol Biomed. 2024 Oct 11;5(1):47. doi: 10.1186/s43556-024-00210-1. Mol Biomed. 2024. PMID: 39390211 Free PMC article. Review.
-
Recent Advances and Future Directions in the Understanding of Mucormycosis.Front Cell Infect Microbiol. 2022 Feb 24;12:850581. doi: 10.3389/fcimb.2022.850581. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35281441 Free PMC article. Review.
-
Myosin-II proteins are involved in the growth, morphogenesis, and virulence of the human pathogenic fungus Mucor circinelloides.Front Cell Infect Microbiol. 2022 Dec 16;12:1031463. doi: 10.3389/fcimb.2022.1031463. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36590583 Free PMC article.
-
Preserving Vascular Integrity Protects Mice against Multidrug-Resistant Gram-Negative Bacterial Infection.Antimicrob Agents Chemother. 2020 Jul 22;64(8):e00303-20. doi: 10.1128/AAC.00303-20. Print 2020 Jul 22. Antimicrob Agents Chemother. 2020. PMID: 32393494 Free PMC article.
References
-
- Sugar A. Principles and Practice of Infectious Diseases. Agents of mucormycosis and related species. In: Mandell GL, Bennett JE, Dolin R, eds. Philadelphia, Pennsylvania, USA: Elsevier; 2005:2979.
-
- Ibrahim AS, Edwards JJE, Filler SG, Spellberg B. Essentials of Clinical Mycology. 2011. Mucormycosis and entomophthoramycosis (zygomycosis). pp. 265–280. In: Kauffman CA, Pappas PG, Sobel JD, Dismukes WE, eds. New York, New York, USA: Springer;
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

