Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets
- PMID: 24356096
- PMCID: PMC3946308
- DOI: 10.1158/2159-8290.CD-13-0286
Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets
Abstract
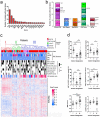
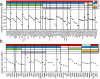
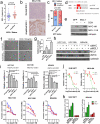
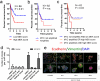
Neoadjuvant chemotherapy (NAC) induces a pathologic complete response (pCR) in approximately 30% of patients with triple-negative breast cancers (TNBC). In patients lacking a pCR, NAC selects a subpopulation of chemotherapy-resistant tumor cells. To understand the molecular underpinnings driving treatment-resistant TNBCs, we performed comprehensive molecular analyses on the residual disease of 74 clinically defined TNBCs after NAC, including next-generation sequencing (NGS) on 20 matched pretreatment biopsies. Combined NGS and digital RNA expression analysis identified diverse molecular lesions and pathway activation in drug-resistant tumor cells. Ninety percent of the tumors contained a genetic alteration potentially treatable with a currently available targeted therapy. Thus, profiling residual TNBCs after NAC identifies targetable molecular lesions in the chemotherapy-resistant component of the tumor, which may mirror micrometastases destined to recur clinically. These data can guide biomarker-driven adjuvant studies targeting these micrometastases to improve the outcome of patients with TNBC who do not respond completely to NAC.
Significance: This study demonstrates the spectrum of genomic alterations present in residual TNBC after NAC. Because TNBCs that do not achieve a CR after NAC are likely to recur as metastatic disease at variable times after surgery, these alterations may guide the selection of targeted therapies immediately after mastectomy before these metastases become evident.
2013 AACR
Figures
References
-
- Guarneri V, Broglio K, Kau SW, Cristofanilli M, Buzdar AU, Valero V, et al. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol. 2006;24:1037–44. - PubMed
-
- Kuerer HM, Newman LA, Smith TL, Ames FC, Hunt KK, Dhingra K, et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol. 1999;17:460–9. - PubMed
-
- Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. - PubMed
-
- Guarneri V, Piacentini F, Ficarra G, Frassoldati A, D'Amico R, Giovannelli S, et al. A prognostic model based on nodal status and Ki-67 predicts the risk of recurrence and death in breast cancer patients with residual disease after preoperative chemotherapy. Ann Oncol. 2009;20:1193–8. - PubMed
-
- Jones RL, Salter J, A'Hern R, Nerurkar A, Parton M, Reis-Filho JS, et al. The prognostic significance of Ki67 before and after neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2009;116:53–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

