Computational assay of H7N9 influenza neuraminidase reveals R292K mutation reduces drug binding affinity
- PMID: 24356381
- PMCID: PMC3868970
- DOI: 10.1038/srep03561
Computational assay of H7N9 influenza neuraminidase reveals R292K mutation reduces drug binding affinity
Abstract
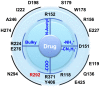
The emergence of a novel H7N9 avian influenza that infects humans is a serious cause for concern. Of the genome sequences of H7N9 neuraminidase available, one contains a substitution of arginine to lysine at position 292, suggesting a potential for reduced drug binding efficacy. We have performed molecular dynamics simulations of oseltamivir, zanamivir and peramivir bound to H7N9, H7N9-R292K, and a structurally related H11N9 neuraminidase. They show that H7N9 neuraminidase is structurally homologous to H11N9, binding the drugs in identical modes. The simulations reveal that the R292K mutation disrupts drug binding in H7N9 in a comparable manner to that observed experimentally for H11N9-R292K. Absolute binding free energy calculations with the WaterSwap method confirm a reduction in binding affinity. This indicates that the efficacy of antiviral drugs against H7N9-R292K will be reduced. Simulations can assist in predicting disruption of binding caused by mutations in neuraminidase, thereby providing a computational 'assay.'
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

