SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements
- PMID: 24356447
- PMCID: PMC3923644
- DOI: 10.1091/mbc.E13-05-0286
SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements
Abstract
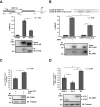
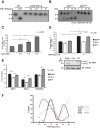
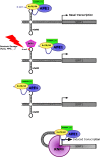
Apurinic/apyrimidinic endonuclease 1 (APE1) is a multifunctional protein contributing to genome stability via repair of DNA lesions via the base excision repair pathway. It also plays a role in gene expression regulation and RNA metabolism. Another, poorly characterized function is its ability to bind to negative calcium responsive elements (nCaRE) of some gene promoters. The presence of many functional nCaRE sequences regulating gene transcription can be envisioned, given their conservation within ALU repeats. To look for functional nCaRE sequences within the human genome, we performed bioinformatic analyses and identified 57 genes potentially regulated by APE1. We focused on sirtuin-1 (SIRT1) deacetylase due to its involvement in cell stress, including senescence, apoptosis, and tumorigenesis, and its role in the deacetylation of APE1 after genotoxic stress. The human SIRT1 promoter presents two nCaRE elements stably bound by APE1 through its N-terminus. We demonstrate that APE1 is part of a multiprotein complex including hOGG1, Ku70, and RNA Pol II, which is recruited on SIRT1 promoter to regulate SIRT1 gene functions during early response to oxidative stress. These findings provide new insights into the role of nCaRE sequences in the transcriptional regulation of mammalian genes.
Figures



References
-
- Alvarez D, Novac O, Callejo M, Ruiz MT, Price GB, Zannis-Hadjopoulos M. 14-3-3sigma is a cruciform DNA binding protein and associates in vivo with origins of DNA replication. J Cell Biochem. 2002;87:194–207. - PubMed
-
- Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

