Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs
- PMID: 24356509
- PMCID: PMC3905700
- DOI: 10.1038/ncomms3980
Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs
Abstract
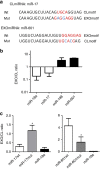
Exosomes are released by most cells to the extracellular environment and are involved in cell-to-cell communication. Exosomes contain specific repertoires of mRNAs, microRNAs (miRNAs) and other non-coding RNAs that can be functionally transferred to recipient cells. However, the mechanisms that control the specific loading of RNA species into exosomes remain unknown. Here we describe sequence motifs present in miRNAs that control their localization into exosomes. The protein heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) specifically binds exosomal miRNAs through the recognition of these motifs and controls their loading into exosomes. Moreover, hnRNPA2B1 in exosomes is sumoylated, and sumoylation controls the binding of hnRNPA2B1 to miRNAs. The loading of miRNAs into exosomes can be modulated by mutagenesis of the identified motifs or changes in hnRNPA2B1 expression levels. These findings identify hnRNPA2B1 as a key player in miRNA sorting into exosomes and provide potential tools for the packaging of selected regulatory RNAs into exosomes and their use in biomedical applications.
Figures



References
-
- Thery C., Ostrowski M. & Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 (2009). - PubMed
-
- Valadi H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 9, 654–659 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

