Functional, anatomical, and molecular investigation of the cardiac conduction system and arrhythmogenic atrioventricular ring tissue in the rat heart
- PMID: 24356527
- PMCID: PMC3886739
- DOI: 10.1161/JAHA.113.000246
Functional, anatomical, and molecular investigation of the cardiac conduction system and arrhythmogenic atrioventricular ring tissue in the rat heart
Abstract
Background: The cardiac conduction system consists of the sinus node, nodal extensions, atrioventricular (AV) node, penetrating bundle, bundle branches, and Purkinje fibers. Node-like AV ring tissue also exists at the AV junctions, and the right and left rings unite at the retroaortic node. The study aims were to (1) construct a 3-dimensional anatomical model of the AV rings and retroaortic node, (2) map electrical activation in the right ring and study its action potential characteristics, and (3) examine gene expression in the right ring and retroaortic node.
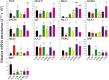
Methods and results: Three-dimensional reconstruction (based on magnetic resonance imaging, histology, and immunohistochemistry) showed the extent and organization of the specialized tissues (eg, how the AV rings form the right and left nodal extensions into the AV node). Multiextracellular electrode array and microelectrode mapping of isolated right ring preparations revealed robust spontaneous activity with characteristic diastolic depolarization. Using laser microdissection gene expression measured at the mRNA level (using quantitative PCR) and protein level (using immunohistochemistry and Western blotting) showed that the right ring and retroaortic node, like the sinus node and AV node but, unlike ventricular muscle, had statistically significant higher expression of key transcription factors (including Tbx3, Msx2, and Id2) and ion channels (including HCN4, Cav3.1, Cav3.2, Kv1.5, SK1, Kir3.1, and Kir3.4) and lower expression of other key ion channels (Nav1.5 and Kir2.1).
Conclusions: The AV rings and retroaortic node possess gene expression profiles similar to that of the AV node. Ion channel expression and electrophysiological recordings show the AV rings could act as ectopic pacemakers and a source of atrial tachycardia.
Keywords: action potential; arrhythmogenesis; atrioventricular ring tissues; cardiac conduction system; ion channels.
Figures

















References
-
- Anderson RH, Yanni J, Boyett MR, Chandler NJ, Dobrzynski H. The anatomy of the cardiac conduction system. Clin Anat. 2009; 22:99-113 - PubMed
-
- Yanni J, Boyett MR, Anderson RH, Dobrzynski H. The extent of the specialized atrioventricular ring tissues. Heart Rhythm. 2009; 6:672-680 - PubMed
-
- Kistler PM, Roberts‐Thomson KC, Haqqani HM, Fynn SP, Singarayar S, Vohra JK, Morton JB, Sparks PB, Kalman JM. P‐wave morphology in focal atrial tachycardia: development of an algorithm to predict the anatomic site of origin. J Am Coll Cardiol. 2006; 48:1010-1017 - PubMed
-
- Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004; 62:489-499 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

