Using Spinach-based sensors for fluorescence imaging of intracellular metabolites and proteins in living bacteria
- PMID: 24356773
- PMCID: PMC4028027
- DOI: 10.1038/nprot.2014.001
Using Spinach-based sensors for fluorescence imaging of intracellular metabolites and proteins in living bacteria
Abstract
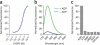
Genetically encoded fluorescent sensors can be valuable tools for studying the abundance and flux of molecules in living cells. We recently developed a novel class of sensors composed of RNAs that can be used to detect diverse small molecules and untagged proteins. These sensors are based on Spinach, an RNA mimic of GFP, and they have successfully been used to image several metabolites and proteins in living bacteria. Here we discuss the generation and optimization of these Spinach-based sensors, which, unlike most currently available genetically encoded reporters, can be readily generated to any target of interest. We also provide a detailed protocol for imaging ADP dynamics in living Escherichia coli after a change from glucose-containing medium to other carbon sources. The entire procedure typically takes ∼4 d including bacteria transformation and image analysis. The majority of this protocol is applicable to sensing other metabolites and proteins in living bacteria.
Figures
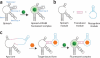



References
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

