Glioblastoma cells containing mutations in the cohesin component STAG2 are sensitive to PARP inhibition
- PMID: 24356817
- PMCID: PMC4130349
- DOI: 10.1158/1535-7163.MCT-13-0749
Glioblastoma cells containing mutations in the cohesin component STAG2 are sensitive to PARP inhibition
Abstract
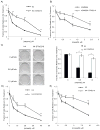
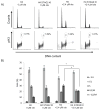
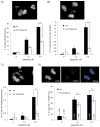
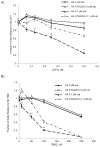
Recent data have identified STAG2, a core subunit of the multifunctional cohesin complex, as a highly recurrently mutated gene in several types of cancer. We sought to identify a therapeutic strategy to selectively target cancer cells harboring inactivating mutations of STAG2 using two independent pairs of isogenic glioblastoma cell lines containing either an endogenous mutant STAG2 allele or a wild-type STAG2 allele restored by homologous recombination. We find that mutations in STAG2 are associated with significantly increased sensitivity to inhibitors of the DNA repair enzyme PARP. STAG2-mutated, PARP-inhibited cells accumulated in G2 phase and had a higher percentage of micronuclei, fragmented nuclei, and chromatin bridges compared with wild-type STAG2 cells. We also observed more 53BP1 foci in STAG2-mutated glioblastoma cells, suggesting that these cells have defects in DNA repair. Furthermore, cells with mutations in STAG2 were more sensitive than cells with wild-type STAG2 when PARP inhibitors were used in combination with DNA-damaging agents. These data suggest that PARP is a potential target for tumors harboring inactivating mutations in STAG2, and strongly recommend that STAG2 status be determined and correlated with therapeutic response to PARP inhibitors, both prospectively and retrospectively, in clinical trials.
©2013 AACR.
Conflict of interest statement
The authors disclose no potential conflicts of interest
Figures
References
-
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. - PubMed
-
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. - PubMed
-
- Gottipati P, Vischioni B, Schultz N, Solomons J, Bryant HE, Djureinovic T, et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010;70:5389–98. - PubMed
-
- McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

