Characterization and structure of the vaccinia virus NF-κB antagonist A46
- PMID: 24356965
- PMCID: PMC3916572
- DOI: 10.1074/jbc.M113.512756
Characterization and structure of the vaccinia virus NF-κB antagonist A46
Abstract
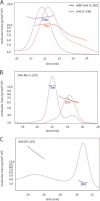
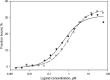
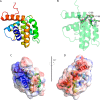
Successful vaccinia virus (VACV) replication in the host requires expression of viral proteins that interfere with host immunity, such as antagonists of the activation of the proinflammatory transcription factor NF-κB. Two such VACV proteins are A46 and A52. A46 interacts with the Toll-like receptor/interleukin-1R (TIR) domain of Toll-like receptors and intracellular adaptors such as MAL (MyD88 adapter-like), TRAM (TIR domain-containing adapter-inducing interferon-β (TRIF)-related adaptor molecule), TRIF, and MyD88, whereas A52 binds to the downstream signaling components TRAF6 and IRAK2. Here, we characterize A46 biochemically, determine by microscale thermophoresis binding constants for the interaction of A46 with the TIR domains of MyD88 and MAL, and present the 2.0 Å resolution crystal structure of A46 residues 87-229. Full-length A46 behaves as a tetramer; variants lacking the N-terminal 80 residues are dimeric. Nevertheless, both bind to the Toll-like receptor domains of MAL and MyD88 with KD values in the low μm range. Like A52, A46 also shows a Bcl-2-like fold but with biologically relevant differences from that of A52. Thus, A46 uses helices α4 and α6 to dimerize, compared with the α1-α6 face used by A52 and other Bcl-2 like VACV proteins. Furthermore, the loop between A46 helices α4-α5 is flexible and shorter than in A52; there is also evidence for an intramolecular disulfide bridge between consecutive cysteine residues. We used molecular docking to propose how A46 interacts with the BB loop of the TRAM TIR domain. Comparisons of A46 and A52 exemplify how subtle changes in viral proteins with the same fold lead to crucial differences in biological activity.
Keywords: Bcl-2 Family Proteins; Inflammation; MyD88; Protein-Protein Interactions; Viral Immunology.
Figures


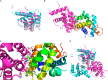

References
-
- Medzhitov R., Janeway C., Jr. (2000) Innate immunity. N. Engl. J. Med. 343, 338–344 - PubMed
-
- Aoshi T., Koyama S., Kobiyama K., Akira S., Ishii K. J. (2011) Innate and adaptive immune responses to viral infection and vaccination. Curr. Opin. Virol. 1, 226–232 - PubMed
-
- O'Neill L. A. (2006) How Toll-like receptors signal. What we know and what we don't know. Curr. Opin. Immunol. 18, 3–9 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

