A novel method for detecting uniparental disomy from trio genotypes identifies a significant excess in children with developmental disorders
- PMID: 24356988
- PMCID: PMC3975066
- DOI: 10.1101/gr.160465.113
A novel method for detecting uniparental disomy from trio genotypes identifies a significant excess in children with developmental disorders
Abstract
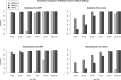
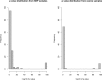
Exome sequencing of parent-offspring trios is a popular strategy for identifying causative genetic variants in children with rare diseases. This method owes its strength to the leveraging of inheritance information, which facilitates de novo variant calling, inference of compound heterozygosity, and the identification of inheritance anomalies. Uniparental disomy describes the inheritance of a homologous chromosome pair from only one parent. This aberration is important to detect in genetic disease studies because it can result in imprinting disorders and recessive diseases. We have developed a software tool to detect uniparental disomy from child-mother-father genotype data that uses a binomial test to identify chromosomes with a significant burden of uniparentally inherited genotypes. This tool is the first to read VCF-formatted genotypes, to perform integrated copy number filtering, and to use a statistical test inherently robust for use in platforms of varying genotyping density and noise characteristics. Simulations demonstrated superior accuracy compared with previously developed approaches. We implemented the method on 1057 trios from the Deciphering Developmental Disorders project, a trio-based rare disease study, and detected six validated events, a significant enrichment compared with the population prevalence of UPD (1 in 3500), suggesting that most of these events are pathogenic. One of these events represents a known imprinting disorder, and exome analyses have identified rare homozygous candidate variants, mainly in the isodisomic regions of UPD chromosomes, which, among other variants, provide targets for further genetic and functional evaluation.
Figures




Similar articles
-
Contribution of uniparental disomy in a clinical trio exome cohort of 2675 patients.Mol Genet Genomic Med. 2021 Nov;9(11):e1792. doi: 10.1002/mgg3.1792. Epub 2021 Sep 29. Mol Genet Genomic Med. 2021. PMID: 34587367 Free PMC article.
-
Accurate detection of clinically relevant uniparental disomy from exome sequencing data.Genet Med. 2020 Apr;22(4):803-808. doi: 10.1038/s41436-019-0704-x. Epub 2019 Nov 26. Genet Med. 2020. PMID: 31767986 Free PMC article.
-
Obesity and developmental delay in a patient with uniparental disomy of chromosome 2.Int J Obes (Lond). 2016 Dec;40(12):1935-1941. doi: 10.1038/ijo.2016.160. Epub 2016 Sep 22. Int J Obes (Lond). 2016. PMID: 27654142
-
Prenatal diagnosis and genetic counseling of uniparental disomy.Taiwan J Obstet Gynecol. 2022 Mar;61(2):210-215. doi: 10.1016/j.tjog.2022.02.006. Taiwan J Obstet Gynecol. 2022. PMID: 35361378 Review.
-
Whole exome sequencing in a patient with uniparental disomy of chromosome 2 and a complex phenotype.Clin Genet. 2013 Sep;84(3):213-22. doi: 10.1111/cge.12064. Epub 2012 Dec 20. Clin Genet. 2013. PMID: 23167750 Free PMC article. Review.
Cited by
-
Chromosomal microarray analysis for prenatal diagnosis of uniparental disomy: a retrospective study.Mol Cytogenet. 2024 Jan 30;17(1):3. doi: 10.1186/s13039-023-00668-8. Mol Cytogenet. 2024. PMID: 38291465 Free PMC article.
-
Uniparental isodisomy caused autosomal recessive diseases: NGS-based analysis allows the concurrent detection of homogenous variants and copy-neutral loss of heterozygosity.Mol Genet Genomic Med. 2019 Oct;7(10):e00945. doi: 10.1002/mgg3.945. Epub 2019 Aug 27. Mol Genet Genomic Med. 2019. PMID: 31454184 Free PMC article.
-
Detection of structural mosaicism from targeted and whole-genome sequencing data.Genome Res. 2017 Oct;27(10):1704-1714. doi: 10.1101/gr.212373.116. Epub 2017 Aug 30. Genome Res. 2017. PMID: 28855261 Free PMC article.
-
Uniparental disomy screen of Irish rare disorder cohort unmasks homozygous variants of clinical significance in the TMCO1 and PRKRA genes.Front Genet. 2022 Sep 14;13:945296. doi: 10.3389/fgene.2022.945296. eCollection 2022. Front Genet. 2022. PMID: 36186440 Free PMC article.
-
Estimation of intrafamilial DNA contamination in family trio genome sequencing using deviation from Mendelian inheritance.Genome Res. 2022 Nov-Dec;32(11-12):2134-2144. doi: 10.1101/gr.276794.122. Epub 2022 Dec 6. Genome Res. 2022. PMID: 36617634 Free PMC article.
References
-
- Astle W, Balding DJ 2009. Population structure and cryptic relatedness in genetic association studies. Stat Sci 24: 451–471
-
- Balciuniene J, Dahl N, Jalonen P, Verhoeven K, Van Camp G, Borg E, Pettersson U, Jazin EE 1999. α-tectorin involvement in hearing disabilities: One gene–two phenotypes. Hum Genet 105: 211–216 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
