The mechanism of translation initiation on Type 1 picornavirus IRESs
- PMID: 24357634
- PMCID: PMC3990684
- DOI: 10.1002/embj.201386124
The mechanism of translation initiation on Type 1 picornavirus IRESs
Abstract
Picornavirus Type 1 IRESs comprise five principal domains (dII-dVI). Whereas dV binds eIF4G, a conserved AUG in dVI was suggested to stimulate attachment of 43S ribosomal preinitiation complexes, which then scan to the initiation codon. Initiation on Type 1 IRESs also requires IRES trans-acting factors (ITAFs), and several candidates have been proposed. Here, we report the in vitro reconstitution of initiation on three Type 1 IRESs: poliovirus (PV), enterovirus 71 (EV71), and bovine enterovirus (BEV). All of them require eIF2, eIF3, eIF4A, eIF4G, eIF4B, eIF1A, and a single ITAF, poly(C) binding protein 2 (PCBP2). In each instance, initiation starts with binding of eIF4G/eIF4A. Subsequent recruitment of 43S complexes strictly requires direct interaction of their eIF3 constituent with eIF4G. The following events can differ between IRESs, depending on the stability of dVI. If it is unstructured (BEV), all ribosomes scan through dVI to the initiation codon, requiring eIF1 to bypass its AUG. If it is structured (PV, EV71), most initiation events occur without inspection of dVI, implying that its AUG does not determine ribosomal attachment.
Figures




Schematic representation of PCBP2 showing the positions of the three KH domains (upper panel). Ribbon diagrams of PCBP2 KH1-KH2 and KH3 domains (PDB: 2JZX and 2P2R) with the linker between them represented by a dashed line (lower panel). Spheres indicate native (C54, C109, C118, C158, C163, C207, C302) and introduced (C308, C330) cysteines. Cysteines that induced cleavage are colored yellow.
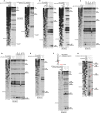
Primer extension analysis after HRC of PV (left) and EV71 (right) IRESs from Fe(II)-tethered PCBP2.
Models of the secondary structures of apical subdomains IVa-IVd of PV and EV71 IRESs, marked to show sites of HRC from PCBP2. Arrow size is proportional to cleavage intensity. Sites of HRC (B, C) are colored to match domains in (A).



References
-
- Anderson EC, Hunt SL, Jackson RJ. Internal initiation of translation from the human rhinovirus-2 internal ribosome entry site requires the binding of Unr to two distinct sites on the 5′ untranslated region. J Gen Virol. 2007;88:3043–3052. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

