Cytosolic cleaved PINK1 represses Parkin translocation to mitochondria and mitophagy
- PMID: 24357652
- PMCID: PMC4303452
- DOI: 10.1002/embr.201337294
Cytosolic cleaved PINK1 represses Parkin translocation to mitochondria and mitophagy
Abstract
PINK1 is a mitochondrial kinase proposed to have a role in the pathogenesis of Parkinson's disease through the regulation of mitophagy. Here, we show that the PINK1 main cleavage product, PINK152, after being generated inside mitochondria, can exit these organelles and localize to the cytosol, where it is not only destined for degradation by the proteasome but binds to Parkin. The interaction of cytosolic PINK1 with Parkin represses Parkin translocation to the mitochondria and subsequent mitophagy. Our work therefore highlights the existence of two cellular pools of PINK1 that have different effects on Parkin translocation and mitophagy.
Figures
PINK1 immunoblot of cytoplasmic and mitochondrial fractions from HeLa cells transfected with PINK1 after 8 h exposure to either DMSO, valinomycin, MG132 or both. TIM23 = mitochondrial marker.
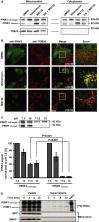
Representative images of PINK1-transfected HeLa cells after 2 h valinomycin or MG132. TOM20 = mitochondrial marker. c = co-localization coefficient. Scale bar = 20 μm.
Alkaline extraction of PINK1 from isolated mitochondria. Upper panel: Representative immunoblot. Lower panel: Quantification of PINK1 in the particulate fraction (not extractable portion) at varying pH. There is a significant interaction between PINK1 fragments and pH (2W-ANOVA, F3,16 = 253.11, P < 0.001). Newman–Keuls test indicates that PINK1 full length is significantly less extractable than PINK1 cleaved at both pH 11.5 and 12. Values are means ± s.e.m. of three independent experiments.
Autoradiogram of in vitro export assay of [35S]-labeled PINK1 in mitochondria and supernatants (see Materials and Methods). Immunoblots for mitochondrial markers MPP and SMAC.

Co-immunoprecipitation assay on lysates of HEK293T cells transfected with C-terminal Flag-tagged human PINK1 or PINK1 fragment constructs (schematic) with an anti-Flag antibody, followed by immunoblotting for Flag and Parkin.
Co-immunoprecipitation assay on lysates of HEK293T cells co-transfected with PINK1 kinase domain (PINK1156–507-HA) and selected myc-tagged Parkin domain constructs (schematic).
PINK1Δ1–103 (used to mimic cleaved PINK1) binds to YFP-Parkin. Co-immunoprecipitation assay on YFP-Parkin HeLa cells transiently transfected or not with PINK1Δ1–103. Rabbit IgG = nonspecific binding control; *Parkin high-molecular weight species.

A, B YFP-Parkin HeLa cells were pretreated with MG132 (5 h) to increase endogenous cleaved PINK1 and then exposed to valinomycin/MG132 for: (A) Parkin translocation (2 h); (B) mitophagy (19 h). Mitophagy was inferred by the disappearance of the mitochondrial network (see representative images in supplementary Fig S3). In (A), there is a significant interaction between membrane potential and proteasome inhibition (2W-ANOVA,F[1,8] = 16.12, P < 0.001). Newman–Keuls test indicates that the number of cells with mitochondrial YFP-Parkin is significantly higher in the DMSO/valinomycin than in the MG132/valinomycin group. In (B), there is a significant interaction among membrane potential, proteasome inhibition, and mitochondrial network (3W-ANOVA,F[2,24] = 239.01, P < 0.001). Newman–Keuls test indicates that the number of cells with normal mitochondrial network is significantly lower in the DMSO/valinomycin than in the MG132/valinomycin group. It also shows that the number of cells with reduced mitochondrial network or no mitochondrial network is significantly higher in the DMSO/valinomycin than in the MG132/valinomycin group.
C, D YFP-Parkin HeLa cells were either transfected with full-length (FL), cleaved mimicking (Δ1–103) PINK1 or not transfected (N-Trans.) and then exposed to valinomycin (C: Parkin translocation [1.5 h]; D: mitophagy [16 h]). In (C), there is a significant interaction between membrane potential and PINK1 fragment (2W-ANOVA,F[2,12] = 68.4, P < 0.001). Newman–Keuls test indicates that the number of cells with mitochondrial YFP-Parkin is significantly higher in N-trans cells than cells expressing PINK1 Δ1–103 after valinomycin treatment. In (D), there is a significant interaction among membrane potential, PINK1 fragment, and mitochondrial network (3W-ANOVA,F[4,36] = 96.01, P < 0.001). Newman–Keuls test indicates that, after valinomycin treatment, the number of cells without mitochondrial network is significantly higher in N-Trans. cells than in cells expressing PINK1 Δ1–103.

Schematic representation of PINK1. MTS: mitochondrial targeting sequence; TM: transmembrane domain; Kinase: kinase domain.
Proposed life cycle of PINK1.
Comment in
-
The many faces of mitophagy.EMBO Rep. 2014 Jan;15(1):5-6. doi: 10.1002/embr.201338224. EMBO Rep. 2014. PMID: 24398127 Free PMC article.
References
-
- Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron. 2003;39:889–909. - PubMed
-
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RLA, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrané J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

