An adaptive coarse graining method for signal transduction in three dimensions
- PMID: 24357890
- PMCID: PMC3865981
- DOI: 10.3233/FI-2012-720
An adaptive coarse graining method for signal transduction in three dimensions
Abstract
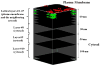
The spatio-temporal landscape of the plasma membrane regulates activation and signal transduction of membrane bound receptors by restricting their two-dimensional mobility and by inducing receptor clustering. This regulation also extends to complex formation between receptors and adaptor proteins, which are the intermediate signaling molecules involved in cellular signaling that relay the received cues from cell surface to cytoplasm and eventually to the nucleus. Although their investigation poses challenging technical difficulties, there is a crucial need to understand the impact of the receptor diffusivity, clustering, and spatial heterogeneity, and of receptor-adaptor protein complex formation on the cellular signal transduction patterns. Building upon our earlier studies, we have developed an adaptive coarse-grained Monte Carlo method that can be used to investigate the role of diffusion, clustering and membrane corralling on receptor association and receptor-adaptor protein complex formation dynamics in three dimensions. The new Monte Carlo lattice based approach allowed us to introduce spatial resolution on the 2-D plasma membrane and to model the cytoplasm in three-dimensions. Being a multi-resolution approach, our new method makes it possible to represent various parts of the cellular system at different levels of detail and enabled us to utilize the locally homogeneous assumption when justified (e.g., cytoplasmic region away from the cell membrane) and avoid its use when high spatial resolution is needed (e.g., cell membrane and cytoplasmic region near the membrane) while keeping the required computational complexity manageable. Our results have shown that diffusion has a significant impact on receptor-receptor dimerization and receptor-adaptor protein complex formation kinetics. We have observed an "adaptor protein hopping" mechanism where the receptor binding proteins may hop between receptors to form short-lived transient complexes. This increased residence time of the adaptor proteins near cell membrane and their ability to frequently change signaling partners may explain the increase in signaling efficiency when receptors are clustered. We also hypothesize that the adaptor protein hopping mechanism can cause concurrent or sequential activation of multiple signaling pathways, thus leading to crosstalk between diverse biological functions.
Figures



References
-
- Berg HC. Random walks in biology. Princeton, NJ: Princeton University Press; 1993.
-
- Carpenter S, O’Neill LA. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem J. 2009;422(1):1–10. - PubMed
-
- Chatterjee A, Katsoulakis MA, et al. Spatially adaptive grand canonical ensemble Monte Carlo simulations. Physical Review E. 2005;71(2) - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
