A genome-wide search for linkage of estimated glomerular filtration rate (eGFR) in the Family Investigation of Nephropathy and Diabetes (FIND)
- PMID: 24358131
- PMCID: PMC3866106
- DOI: 10.1371/journal.pone.0081888
A genome-wide search for linkage of estimated glomerular filtration rate (eGFR) in the Family Investigation of Nephropathy and Diabetes (FIND)
Abstract
Objective: Estimated glomerular filtration rate (eGFR), a measure of kidney function, is heritable, suggesting that genes influence renal function. Genes that influence eGFR have been identified through genome-wide association studies. However, family-based linkage approaches may identify loci that explain a larger proportion of the heritability. This study used genome-wide linkage and association scans to identify quantitative trait loci (QTL) that influence eGFR.
Methods: Genome-wide linkage and sparse association scans of eGFR were performed in families ascertained by probands with advanced diabetic nephropathy (DN) from the multi-ethnic Family Investigation of Nephropathy and Diabetes (FIND) study. This study included 954 African Americans (AA), 781 American Indians (AI), 614 European Americans (EA) and 1,611 Mexican Americans (MA). A total of 3,960 FIND participants were genotyped for 6,000 single nucleotide polymorphisms (SNPs) using the Illumina Linkage IVb panel. GFR was estimated by the Modification of Diet in Renal Disease (MDRD) formula.
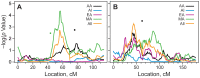
Results: The non-parametric linkage analysis, accounting for the effects of diabetes duration and BMI, identified the strongest evidence for linkage of eGFR on chromosome 20q11 (log of the odds [LOD] = 3.34; P = 4.4 × 10(-5)) in MA and chromosome 15q12 (LOD = 2.84; P = 1.5 × 10(-4)) in EA. In all subjects, the strongest linkage signal for eGFR was detected on chromosome 10p12 (P = 5.5 × 10(-4)) at 44 cM near marker rs1339048. A subsequent association scan in both ancestry-specific groups and the entire population identified several SNPs significantly associated with eGFR across the genome.
Conclusion: The present study describes the localization of QTL influencing eGFR on 20q11 in MA, 15q21 in EA and 10p12 in the combined ethnic groups participating in the FIND study. Identification of causal genes/variants influencing eGFR, within these linkage and association loci, will open new avenues for functional analyses and development of novel diagnostic markers for DN.
Conflict of interest statement
Figures

References
-
- Rossing P, de Zeeuw D (2011) Need for better diabetes treatment for improved renal outcome. Kidney Int Suppl 120: S28–S32. - PubMed
-
- Nelson RG, Pettitt DJ, Carraher MJ, Baird HR, Knowler WC (1998) Effect of proteinuria on mortality in NIDDM. Diabetes 37: 1499–504. - PubMed
-
- Hunt SC, Coon H, Hasstedt SJ, Cawthon RM, Camp NJ, et al. (2004) Linkage of serum creatinine and glomerular filtration rate to chromosome 2 in Utah pedigrees. Am J Hypertens 17: 511–515. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

