Microbiota and metabolite profiling reveal specific alterations in bacterial community structure and environment in the cystic fibrosis airway during exacerbation
- PMID: 24358183
- PMCID: PMC3866110
- DOI: 10.1371/journal.pone.0082432
Microbiota and metabolite profiling reveal specific alterations in bacterial community structure and environment in the cystic fibrosis airway during exacerbation
Abstract
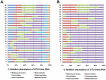
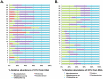
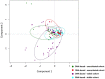
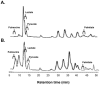
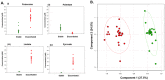
Chronic polymicrobial infections of the lung are the foremost cause of morbidity and mortality in cystic fibrosis (CF) patients. The composition of the microbial flora of the airway alters considerably during infection, particularly during patient exacerbation. An understanding of which organisms are growing, their environment and their behaviour in the airway is of importance for designing antibiotic treatment regimes and for patient prognosis. To this end, we have analysed sputum samples taken from separate cohorts of CF and non-CF subjects for metabolites and in parallel, and we have examined both isolated DNA and RNA for the presence of 16S rRNA genes and transcripts by high-throughput sequencing of amplicon or cDNA libraries. This analysis revealed that although the population size of all dominant orders of bacteria as measured by DNA- and RNA- based methods are similar, greater discrepancies are seen with less prevalent organisms, some of which we associated with CF for the first time. Additionally, we identified a strong relationship between the abundance of specific anaerobes and fluctuations in several metabolites including lactate and putrescine during patient exacerbation. This study has hence identified organisms whose occurrence within the CF microbiome has been hitherto unreported and has revealed potential metabolic biomarkers for exacerbation.
Conflict of interest statement
Figures

References
-
- Rogers GB, Carroll MP, Bruce KD (2009) Studying bacterial infections through culture-independent approaches. J Med Microbiol 58: 1401–1418. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

