Comprehensive analysis of the overall codon usage patterns in equine infectious anemia virus
- PMID: 24359511
- PMCID: PMC3878193
- DOI: 10.1186/1743-422X-10-356
Comprehensive analysis of the overall codon usage patterns in equine infectious anemia virus
Abstract
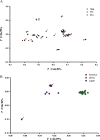
Background: Equine infectious anemia virus (EIAV) is an important animal model for understanding the relationship between viral persistence and the host immune response during lentiviral infections. Comparison and analysis of the codon usage model between EIAV and its hosts is important for the comprehension of viral evolution. In our study, the codon usage pattern of EIAV was analyzed from the available 29 full-length EIAV genomes through multivariate statistical methods.
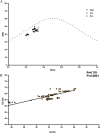
Finding: Effective number of codons (ENC) suggests that the codon usage among EIAV strains is slightly biased. The ENC-plot analysis demonstrates that mutation pressure plays a substantial role in the codon usage pattern of EIAV, whereas other factors such as geographic distribution and host translation selection also take part in the process of EIAV evolution. Comparative analysis of codon adaptation index (CAI) values among EIAV and its hosts suggests that EIAV utilize the translational resources of horse more efficiently than that of donkey.
Conclusion: The codon usage bias in EIAV is slight and mutation pressure is the main factor that affects codon usage variation in EIAV. These results suggest that EIAV genomic biases are the result of the co-evolution of genome composition and the ability to evade the host's immune response.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

