A pipeline of programs for collecting and analyzing group II intron retroelement sequences from GenBank
- PMID: 24359548
- PMCID: PMC4028801
- DOI: 10.1186/1759-8753-4-28
A pipeline of programs for collecting and analyzing group II intron retroelement sequences from GenBank
Abstract
Background: Accurate and complete identification of mobile elements is a challenging task in the current era of sequencing, given their large numbers and frequent truncations. Group II intron retroelements, which consist of a ribozyme and an intron-encoded protein (IEP), are usually identified in bacterial genomes through their IEP; however, the RNA component that defines the intron boundaries is often difficult to identify because of a lack of strong sequence conservation corresponding to the RNA structure. Compounding the problem of boundary definition is the fact that a majority of group II intron copies in bacteria are truncated.
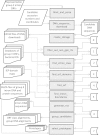
Results: Here we present a pipeline of 11 programs that collect and analyze group II intron sequences from GenBank. The pipeline begins with a BLAST search of GenBank using a set of representative group II IEPs as queries. Subsequent steps download the corresponding genomic sequences and flanks, filter out non-group II introns, assign introns to phylogenetic subclasses, filter out incomplete and/or non-functional introns, and assign IEP sequences and RNA boundaries to the full-length introns. In the final step, the redundancy in the data set is reduced by grouping introns into sets of ≥95% identity, with one example sequence chosen to be the representative.
Conclusions: These programs should be useful for comprehensive identification of group II introns in sequence databases as data continue to rapidly accumulate.
Figures

References
-
- Fedorova O, Zingler N. Group II introns: structure, folding and splicing mechanism. Biol Chem. 2007;4:665–678. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

