Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation
- PMID: 24360274
- PMCID: PMC3905448
- DOI: 10.1016/j.cell.2013.11.039
Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation
Abstract
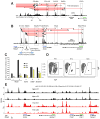
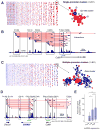
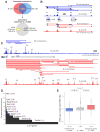
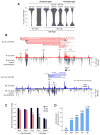
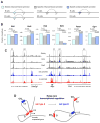
A key finding of the ENCODE project is that the enhancer landscape of mammalian cells undergoes marked alterations during ontogeny. However, the nature and extent of these changes are unclear. As part of the NIH Mouse Regulome Project, we here combined DNaseI hypersensitivity, ChIP-seq, and ChIA-PET technologies to map the promoter-enhancer interactomes of pluripotent ES cells and differentiated B lymphocytes. We confirm that enhancer usage varies widely across tissues. Unexpectedly, we find that this feature extends to broadly transcribed genes, including Myc and Pim1 cell-cycle regulators, which associate with an entirely different set of enhancers in ES and B cells. By means of high-resolution CpG methylomes, genome editing, and digital footprinting, we show that these enhancers recruit lineage-determining factors. Furthermore, we demonstrate that the turning on and off of enhancers during development correlates with promoter activity. We propose that organisms rely on a dynamic enhancer landscape to control basic cellular functions in a tissue-specific manner.
Copyright © 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
J.K.J. has a financial interest in Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Figures


References
-
- Casellas R, Jankovic M, Meyer G, Gazumyan A, Luo Y, Roeder R, Nussenzweig M. OcaB is required for normal transcription and V(D)J recombination of a subset of immunoglobulin kappa genes. Cell. 2002;110:575–585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

