Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex
- PMID: 24360784
- PMCID: PMC4349574
- DOI: 10.1016/j.devcel.2013.11.012
Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex
Abstract
Plasma membrane PI4P helps determine the identity of this membrane and plays a key role in signal transduction as the precursor of PI(4,5)P2 and its metabolites. Here, we report the atomic structure of the protein scaffold that is required for the plasma membrane localization and function of Stt4/PI4KIIIα, the PI 4-kinase responsible for this PI4P pool. Both proteins of the scaffold, Efr3 and YPP1/TTC7, are composed of α-helical repeats, which are arranged into a rod in Efr3 and a superhelix in Ypp1. A conserved basic patch in Efr3, which binds acidic phospholipids, anchors the complex to the plasma membrane. Stt4/PI4KIIIα is recruited by interacting with the Ypp1 C-terminal lobe, which also binds to unstructured regions in the Efr3 C terminus. Phosphorylation of this Efr3 region counteracts Ypp1 binding, thus providing a mechanism through which Stt4/PI4KIIIα recruitment, and thus a metabolic reaction of fundamental importance in cell physiology, can be regulated.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
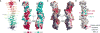




References
-
- Abramoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42.
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

