A UV-induced genetic network links the RSC complex to nucleotide excision repair and shows dose-dependent rewiring
- PMID: 24360959
- PMCID: PMC4136507
- DOI: 10.1016/j.celrep.2013.11.035
A UV-induced genetic network links the RSC complex to nucleotide excision repair and shows dose-dependent rewiring
Abstract
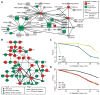
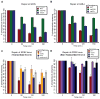
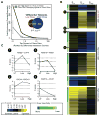
Efficient repair of UV-induced DNA damage requires the precise coordination of nucleotide excision repair (NER) with numerous other biological processes. To map this crosstalk, we generated a differential genetic interaction map centered on quantitative growth measurements of >45,000 double mutants before and after different doses of UV radiation. Integration of genetic data with physical interaction networks identified a global map of 89 UV-induced functional interactions among 62 protein complexes, including a number of links between the RSC complex and several NER factors. We show that RSC is recruited to both silenced and transcribed loci following UV damage where it facilitates efficient repair by promoting nucleosome remodeling. Finally, a comparison of the response to high versus low levels of UV shows that the degree of genetic rewiring correlates with dose of UV and reveals a network of dose-specific interactions. This study makes available a large resource of UV-induced interactions, and it illustrates a methodology for identifying dose-dependent interactions based on quantitative shifts in genetic networks.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Molecular cell. 2004;16:117–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

