Alanine scan of core positions in ubiquitin reveals links between dynamics, stability, and function
- PMID: 24361330
- PMCID: PMC3951674
- DOI: 10.1016/j.jmb.2013.10.042
Alanine scan of core positions in ubiquitin reveals links between dynamics, stability, and function
Abstract
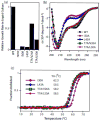
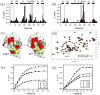
Mutations at solvent-inaccessible core positions in proteins can impact function through many biophysical mechanisms including alterations to thermodynamic stability and protein dynamics. As these properties of proteins are difficult to investigate, the impacts of core mutations on protein function are poorly understood for most systems. Here, we determined the effects of alanine mutations at all 15 core positions in ubiquitin on function in yeast. The majority (13 of 15) of alanine substitutions supported yeast growth as the sole ubiquitin. Both the two null mutants (I30A and L43A) were less stable to temperature-induced unfolding in vitro than wild type (WT) but were well folded at physiological temperatures. Heteronuclear NMR studies indicated that the L43A mutation reduces temperature stability while retaining a ground-state structure similar to WT. This structure enables L43A to bind to common ubiquitin receptors in vitro. Many of the core alanine ubiquitin mutants, including one of the null variants (I30A), exhibited an increased accumulation of high-molecular-weight species, suggesting that these mutants caused a defect in the processing of ubiquitin-substrate conjugates. In contrast, L43A exhibited a unique accumulation pattern with reduced levels of high-molecular-weight species and undetectable levels of free ubiquitin. When conjugation to other proteins was blocked, L43A ubiquitin accumulated as free ubiquitin in yeast. Based on these findings, we speculate that ubiquitin's stability to unfolding may be required for efficient recycling during proteasome-mediated substrate degradation.
Keywords: proteasome; protein function; structural dynamics; thermodynamic stability; ubiquitin recycling.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




Comment on
-
Analyses of the effects of all ubiquitin point mutants on yeast growth rate.J Mol Biol. 2013 Apr 26;425(8):1363-77. doi: 10.1016/j.jmb.2013.01.032. Epub 2013 Jan 30. J Mol Biol. 2013. PMID: 23376099 Free PMC article.
References
-
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–45. - PubMed
-
- Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–11. - PubMed
-
- Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–5. - PubMed
-
- Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

