Heparan sulfate 3-O-sulfation: a rare modification in search of a function
- PMID: 24361527
- PMCID: PMC4039620
- DOI: 10.1016/j.matbio.2013.12.001
Heparan sulfate 3-O-sulfation: a rare modification in search of a function
Abstract
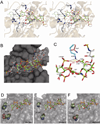
Many protein ligands bind to heparan sulfate, which results in their presentation, protection, oligomerization or conformational activation. Binding depends on the pattern of sulfation and arrangement of uronic acid epimers along the chains. Sulfation at the C3 position of glucosamine is a relatively rare, yet biologically significant modification, initially described as a key determinant for binding and activation of antithrombin and later for infection by type I herpes simplex virus. In mammals, a family of seven heparan sulfate 3-O-sulfotransferases installs sulfate groups at this position and constitutes the largest group of sulfotransferases involved in heparan sulfate formation. However, to date very few proteins or biological systems have been described that are influenced by 3-O-sulfation. This review describes our current understanding of the prevalence and structure of 3-O-sulfation sites, expression and substrate specificity of the 3-O-sulfotransferase family and the emerging roles of 3-O-sulfation in biology.
Keywords: 3-O-Sulfotransferases; Antithrombin; Growth factors; Heparan sulfate; Sulfation.
Copyright © 2013 International Society of Matrix Biology. Published by Elsevier B.V. All rights reserved.
Figures
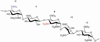


References
-
- Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry. 1985;24:6723–6729. - PubMed
-
- Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of 3-O- and 6-O-sulfated glucosamine residues in the heparin-induced conformational change in antithrombin III. Biochemistry. 1987;26:6454–6461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

