A thermodynamic assay to test pharmacological chaperones for Fabry disease
- PMID: 24361605
- PMCID: PMC3909460
- DOI: 10.1016/j.bbagen.2013.12.018
A thermodynamic assay to test pharmacological chaperones for Fabry disease
Abstract
Background: The majority of the disease-causing mutations affect protein stability, but not functional sites and are amenable, in principle, to be treated with pharmacological chaperones. These drugs enhance the thermodynamic stability of their targets. Fabry disease, a disorder caused by mutations in the gene encoding lysosomal alpha-galactosidase, represents an excellent model system to develop experimental protocols to test the efficiency of such drugs.
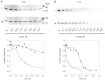
Methods: The stability of lysosomal alpha-galactosidase under different conditions was studied by urea-induced unfolding followed by limited proteolysis and Western blotting.
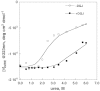
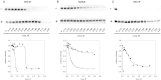
Results: We measured the concentration of urea needed to obtain half-maximal unfolding because this parameter represents an objective indicator of protein stability.
Conclusions: Urea-induced unfolding is a versatile technique that can be adapted to cell extracts containing tiny amounts of wild-type or mutant proteins. It allows testing of protein stability as a function of pH, in the presence or in the absence of drugs. Results are not influenced by the method used to express the protein in transfected cells.
General significance: Scarce and dispersed populations pose a problem for the clinical trial of drugs for rare diseases. This is particularly true for pharmacological chaperones that must be tested on each mutation associated with a given disease. Diverse in vitro tests are needed. We used a method based on chemically induced unfolding as a tool to assess whether a particular Fabry mutation is responsive to pharmacological chaperones, but, by no means is our protocol limited to this disease.
Keywords: 1-deoxy-galactonojirimycin; AGAL; CD; Cell lysate; DGJ; FD; Fabry disease; Limited proteolysis; Lysosomal storage disorder; PC; Pharmacological chaperone; Urea-induced unfolding; circular dichroism; lysosomal alpha-galactosidase; pharmacological chaperones.
Copyright © 2013 The Authors. Published by Elsevier B.V. All rights reserved.
Figures






References
-
- Yue P., Li Z., Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 2005;353:459–473. - PubMed
-
- Boyd R.E., Lee G., Rybczynski P., Benjamin E.R., Khanna R., Wustman B.A., Valenzano K.J. Pharmacological chaperones as therapeutics for lysosomal storage diseases. J. Med. Chem. 2013;56:2705–2725. - PubMed
-
- Young-Gqamana B., Brignol N., Chang H.H., Khanna R., Soska R., Fuller M., Sitaraman S.A., Germain D.P., Giugliani R., Hughes D.A., Mehta A., Nicholls K., Boudes P., Lockhart D.J., Valenzano K.J., Benjamin E.R. Migalastat HCl reduces globotriaosylsphingosine (lyso-Gb3) in Fabry transgenic mice and in the plasma of Fabry patients. PLoS One. 2013;8:e57631. - PMC - PubMed
-
- Benito J.M., Garcia Fernandez J.M., Ortiz Mellet C. Pharmacological chaperone therapy for Gaucher disease: a patent review. Expert. Opin. Ther. Pat. 2011;21:885–903. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

