Non-negative matrix factorization of multimodal MRI, fMRI and phenotypic data reveals differential changes in default mode subnetworks in ADHD
- PMID: 24361664
- PMCID: PMC4063903
- DOI: 10.1016/j.neuroimage.2013.12.015
Non-negative matrix factorization of multimodal MRI, fMRI and phenotypic data reveals differential changes in default mode subnetworks in ADHD
Abstract
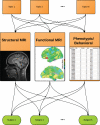
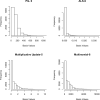
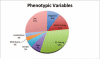
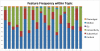
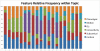
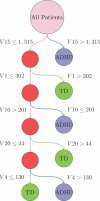
In the multimodal neuroimaging framework, data on a single subject are collected from inherently different sources such as functional MRI, structural MRI, behavioral and/or phenotypic information. The information each source provides is not independent; a subset of features from each modality maps to one or more common latent dimensions, which can be interpreted using generative models. These latent dimensions, or "topics," provide a sparse summary of the generative process behind the features for each individual. Topic modeling, an unsupervised generative model, has been used to map seemingly disparate features to a common domain. We use Non-Negative Matrix Factorization (NMF) to infer the latent structure of multimodal ADHD data containing fMRI, MRI, phenotypic and behavioral measurements. We compare four different NMF algorithms and find that the sparsest decomposition is also the most differentiating between ADHD and healthy patients. We identify dimensions that map to interpretable, recognizable dimensions such as motion, default mode network activity, and other such features of the input data. For example, structural and functional graph theory features related to default mode subnetworks clustered with the ADHD-Inattentive diagnosis. Structural measurements of the default mode network (DMN) regions such as the posterior cingulate, precuneus, and parahippocampal regions were all related to the ADHD-Inattentive diagnosis. Ventral DMN subnetworks may have more functional connections in ADHD-I, while dorsal DMN may have less. ADHD topics are dependent upon diagnostic site, suggesting diagnostic differences across geographic locations. We assess our findings in light of the ADHD-200 classification competition, and contrast our unsupervised, nominated topics with previously published supervised learning methods. Finally, we demonstrate the validity of these latent variables as biomarkers by using them for classification of ADHD in 730 patients. Cumulatively, this manuscript addresses how multimodal data in ADHD can be interpreted by latent dimensions.
Keywords: ADHD; Attention deficit; Biomarkers; Default mode; Latent variables; MRI; Machine learning; Multimodal data; NMF; Phenotype; Sparsity; Topic modeling; fMRI.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

References
-
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders: DSM-IV-TR. American Psychiatric Publishing, Inc.; 2000.
-
- Biswal Bharat, Yetkin F Zerrin, Haughton Victor M, Hyde James S. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic resonance in medicine. 1995;34(4):537–541. - PubMed
-
- Blei David M, Ng Andrew Y, Jordan Michael I. Latent dirichlet allocation. the Journal of machine Learning research. 2003;3:993–1022.
-
- Brown Matthew R G, Sidhu Gagan S, Greiner Russell, Asgarian Nasimeh, Bastani Meysam, Silverstone Peter H, Greenshaw Andrew J, Dursun Serdar M. ADHD-200 global competition: Diagnosing ADHD using personal characteristic data can outperform resting state fMRI measurements. Frontiers in Systems Neuroscience. 2012;6(69) - PMC - PubMed
-
- Buckner Randy L, Andrews-Hanna Jessica R, Schacter Daniel L. The brain's default network. Annals of the New York Academy of Sciences. 2008;1124(1):1–38. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

