Structure of the TbBILBO1 protein N-terminal domain from Trypanosoma brucei reveals an essential requirement for a conserved surface patch
- PMID: 24362019
- PMCID: PMC3916570
- DOI: 10.1074/jbc.M113.529032
Structure of the TbBILBO1 protein N-terminal domain from Trypanosoma brucei reveals an essential requirement for a conserved surface patch
Abstract
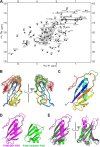
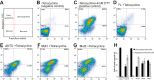
TbBILBO1 is the only known component of the flagellar pocket collar, a cytoskeletal barrier element found in trypanosomes. The N-terminal domain (NTD) of TbBILBO1 was found to be dispensable for targeting of the protein in vivo. However, overexpression of constructs lacking the NTD caused complete growth inhibition, implying an essential requirement for this domain. A high resolution structure of the NTD of TbBILBO1 showed that it forms a ubiquitin-like fold with a conserved surface patch. Mutagenesis of this patch recapitulated the phenotypic effects of deleting the entire domain and was found to cause cell death. The surface patch on the NTD of TbBILBO1 is therefore a potential drug target.
Keywords: Cytoskeleton; Flagellar Pocket Collar; Infectious Diseases; Molecular Cell Biology; Nuclear Magnetic Resonance; Parasite; Protein Structure; Structural Biology; TbBILBO1; Trypanosoma brucei.
Figures






Similar articles
-
The Trypanosome Flagellar Pocket Collar and Its Ring Forming Protein-TbBILBO1.Cells. 2016 Mar 2;5(1):9. doi: 10.3390/cells5010009. Cells. 2016. PMID: 26950156 Free PMC article. Review.
-
Crystal structure of the N-terminal domain of the trypanosome flagellar protein BILBO1 reveals a ubiquitin fold with a long structured loop for protein binding.J Biol Chem. 2020 Feb 7;295(6):1489-1499. doi: 10.1074/jbc.RA119.010768. Epub 2019 Dec 27. J Biol Chem. 2020. PMID: 31882537 Free PMC article.
-
Assembly mechanism of Trypanosoma brucei BILBO1, a multidomain cytoskeletal protein.J Biol Chem. 2014 Aug 22;289(34):23870-81. doi: 10.1074/jbc.M114.554659. Epub 2014 Jul 15. J Biol Chem. 2014. PMID: 25031322 Free PMC article.
-
Expression, purification and preliminary crystallographic analysis of the N-terminal domain of Trypanosoma brucei BILBO1.Acta Crystallogr F Struct Biol Commun. 2014 May;70(Pt 5):628-31. doi: 10.1107/S2053230X14005743. Epub 2014 Apr 17. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 24817725 Free PMC article.
-
Trypanosome glycosylphosphatidylinositol biosynthesis.Korean J Parasitol. 2009 Sep;47(3):197-204. doi: 10.3347/kjp.2009.47.3.197. Epub 2009 Aug 28. Korean J Parasitol. 2009. PMID: 19724691 Free PMC article. Review.
Cited by
-
Intrabody-Induced Cell Death by Targeting the T. brucei Cytoskeletal Protein TbBILBO1.Microbiol Spectr. 2021 Oct 31;9(2):e0091521. doi: 10.1128/Spectrum.00915-21. Epub 2021 Oct 27. Microbiol Spectr. 2021. PMID: 34704826 Free PMC article.
-
Assembly mechanism of Trypanosoma brucei BILBO1 at the flagellar pocket collar.Commun Integr Biol. 2015 Jan 28;8(1):e992739. doi: 10.4161/19420889.2014.992739. eCollection 2015 Jan-Feb. Commun Integr Biol. 2015. PMID: 26844754 Free PMC article.
-
The Trypanosome Flagellar Pocket Collar and Its Ring Forming Protein-TbBILBO1.Cells. 2016 Mar 2;5(1):9. doi: 10.3390/cells5010009. Cells. 2016. PMID: 26950156 Free PMC article. Review.
-
A MORN Repeat Protein Facilitates Protein Entry into the Flagellar Pocket of Trypanosoma brucei.Eukaryot Cell. 2015 Nov;14(11):1081-93. doi: 10.1128/EC.00094-15. Epub 2015 Aug 28. Eukaryot Cell. 2015. PMID: 26318396 Free PMC article.
-
Interaction between the flagellar pocket collar and the hook complex via a novel microtubule-binding protein in Trypanosoma brucei.PLoS Pathog. 2017 Nov 1;13(11):e1006710. doi: 10.1371/journal.ppat.1006710. eCollection 2017 Nov. PLoS Pathog. 2017. PMID: 29091964 Free PMC article.
References
-
- Simpson A. G., Stevens J. R., Lukes J. (2006) The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 22, 168–174 - PubMed
-
- Welburn S. C., Maudlin I. (2012) Priorities for the elimination of sleeping sickness. Adv. Parasitol. 79, 299–337 - PubMed
-
- Schmunis G. A., Yadon Z. E. (2010) Chagas disease: a Latin American health problem becoming a world health problem. Acta Trop. 115, 14–21 - PubMed
-
- Zucca M., Scutera S., Savoia D. (2013) New chemotherapeutic strategies against malaria, leishmaniasis and trypanosomiases. Curr. Med. Chem. 20, 502–526 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

