Discovery and characterization of a new family of lytic polysaccharide monooxygenases
- PMID: 24362702
- PMCID: PMC4274766
- DOI: 10.1038/nchembio.1417
Discovery and characterization of a new family of lytic polysaccharide monooxygenases
Abstract
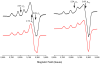
Lytic polysaccharide monooxygenases (LPMOs) are a recently discovered class of enzymes capable of oxidizing recalcitrant polysaccharides. They are attracting considerable attention owing to their potential use in biomass conversion, notably in the production of biofuels. Previous studies have identified two discrete sequence-based families of these enzymes termed AA9 (formerly GH61) and AA10 (formerly CBM33). Here, we report the discovery of a third family of LPMOs. Using a chitin-degrading exemplar from Aspergillus oryzae, we show that the three-dimensional structure of the enzyme shares some features of the previous two classes of LPMOs, including a copper active center featuring the 'histidine brace' active site, but is distinct in terms of its active site details and its EPR spectroscopy. The newly characterized AA11 family expands the LPMO clan, potentially broadening both the range of potential substrates and the types of reactive copper-oxygen species formed at the active site of LPMOs.
Figures


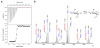
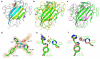
References
-
- Gelfand I, et al. Sustainable bioenergy production from marginal lands in the US Midwest. Nature. 2013;493:514–517. - PubMed
-
- Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330:219–222. - PubMed
-
- Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose Dehydrogenase and a Copper-Dependent Polysaccharide Monooxygenase Potentiate Cellulose Degradation by Neurospora crassa. ACS Chemical Biology. 2011;6:1399–1406. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

