Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2
- PMID: 24363272
- PMCID: PMC3983684
- DOI: 10.1002/embr.201337642
Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2
Abstract
Hypoxia is central to both ischaemic and neoplastic diseases. However, the non-coding transcriptional response to hypoxia is largely uncharacterized. We undertook integrated genomic analyses of both non-coding and coding transcripts using massively parallel sequencing and interfaced this data with pan-genomic analyses of hypoxia-inducible factor (HIF) and RNApol2 binding in hypoxic cells. These analyses revealed that all classes of RNA are profoundly regulated by hypoxia and implicated HIF as a major direct regulator of both the non-coding and coding transcriptome, acting predominantly through release of pre-bound promoter-paused RNApol2. These findings indicate that the transcriptional response to hypoxia is substantially more extensive than previously considered.
Figures

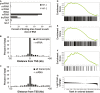
RNA-seq was performed following 24 h culture in 21% (normoxia) or 1% (hypoxia) ambient oxygen.
Outline of the pipeline for mapping ribosomal depleted directional RNA-seq reads.
Raw data plotted as log2 fold-change by hypoxia on the vertical axis versus expression level on the horizontal axis. Vertical lines denote thresholds (higher for classes of longer RNAs) to select transcripts for further analysis.
Box-and-whisker plots of log2 fold-change by hypoxia for filtered transcripts in each class of RNA. The vertical dotted line denotes no fold-change.
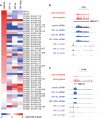
A–C she proportion of HIF-1α and HIF-2α binding sites that are closest to transcribed loci of each RNA class (A). The distribution of (B) HIF-1α and (C) HIF-2α binding sites around the transcriptional start site of the nearest expressed gene irrespective of class (black bars). The grey bars show the distribution when only active mRNA genes are used (analogous to previous analyses).
D–F GSEA analysis against fold-regulation by hypoxia for (D) all HIF-binding transcripts, (E) mRNA and (F) lncRNA.
G Log2 fold-change of transcript abundance in hypoxia, ranked in order from highly upregulated to downregulated transcripts, from which the GSEA were derived.
A Heat map showing fold regulation by hypoxia and by the indicated HIF siRNA for lncRNAs adjacent to HIF-binding site that are detected in the polyA RNA-seq analyses.
B, C RNA-seq and HIF ChIP-seq genome browser tracks for the two most hypoxically upregulated HIF-binding lncRNAs: (B) MALAT1 and (C) NEAT1.
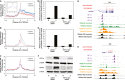
A–C Mean distribution of normoxic (red) and hypoxic (blue) RNApol2 binding at the 100 transcripts most upregulated by hypoxia. Inset shows expanded view at the TSS (FPKM = fragments per kilobase per million reads). The same plots for (B) H3K4me3 and (C) DNAse1 hypersensitivity are shown.
D, E ChIP-qPCR analysis of RNApol2 within the body of (D) ALDOA and (E) NDRG1 gene shows suppression of the hypoxic induction by HIF-1α & HIF-2α siRNA (biological duplicates).
F Immunoblot analysis of HIF-α levels.
G,H RNA-seq and ChIP-seq tracks illustrating (G) release of promoter-paused RNApol2 and (H) de novo recruitment of RNApol2, despite constitutive DNAse1 hypersensitivity.
References
-
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. - PubMed
-
- Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010;38:2332–2345. - PMC - PubMed
-
- McCormick R, Buffa FM, Ragoussis J, Harris AL. The role of hypoxia regulated microRNAs in cancer. Curr Top Microbiol Immunol. 2010;345:47–70. - PubMed
-
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

