Signaling regulates activity of DHCR24, the final enzyme in cholesterol synthesis
- PMID: 24363437
- PMCID: PMC3934726
- DOI: 10.1194/jlr.M043257
Signaling regulates activity of DHCR24, the final enzyme in cholesterol synthesis
Abstract
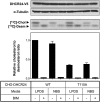
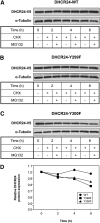
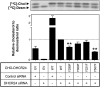
The role of signaling in regulating cholesterol homeostasis is gradually becoming more widely recognized. Here, we explored how kinases and phosphorylation sites regulate the activity of the enzyme involved in the final step of cholesterol synthesis, 3β-hydroxysterol Δ24-reductase (DHCR24). Many factors are known to regulate DHCR24 transcriptionally, but little is known about its posttranslational regulation. We developed a system to specifically test human ectopic DHCR24 activity in a model cell-line (Chinese hamster ovary-7) using siRNA targeted only to hamster DHCR24, thus ensuring that all activity could be attributed to the human enzyme. We determined the effect of known phosphorylation sites and found that mutating certain residues (T110, Y299, and Y507) inhibited DHCR24 activity. In addition, inhibitors of protein kinase C ablated DHCR24 activity, although not through a known phosphorylation site. Our data indicate a novel mechanism whereby DHCR24 activity is regulated by signaling.
Keywords: 3β-hydroxysterol Δ24-reductase; bisindolylmaleimide I; desmosterol; gas chromatography-mass spectrometry; phosphorylation; protein kinase C; regulation.
Figures




Similar articles
-
The endogenous regulator 24(S),25-epoxycholesterol inhibits cholesterol synthesis at DHCR24 (Seladin-1).Biochim Biophys Acta. 2012 Sep;1821(9):1269-77. doi: 10.1016/j.bbalip.2011.11.009. Epub 2011 Dec 10. Biochim Biophys Acta. 2012. PMID: 22178193
-
Desmosterol and DHCR24: unexpected new directions for a terminal step in cholesterol synthesis.Prog Lipid Res. 2013 Oct;52(4):666-80. doi: 10.1016/j.plipres.2013.09.002. Epub 2013 Oct 2. Prog Lipid Res. 2013. PMID: 24095826 Review.
-
The terminal enzymes of cholesterol synthesis, DHCR24 and DHCR7, interact physically and functionally.J Lipid Res. 2015 Apr;56(4):888-97. doi: 10.1194/jlr.M056986. Epub 2015 Jan 31. J Lipid Res. 2015. PMID: 25637936 Free PMC article.
-
Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis.Am J Hum Genet. 2001 Oct;69(4):685-94. doi: 10.1086/323473. Epub 2001 Aug 22. Am J Hum Genet. 2001. PMID: 11519011 Free PMC article.
-
The role of DHCR24 in the pathogenesis of AD: re-cognition of the relationship between cholesterol and AD pathogenesis.Acta Neuropathol Commun. 2022 Mar 16;10(1):35. doi: 10.1186/s40478-022-01338-3. Acta Neuropathol Commun. 2022. PMID: 35296367 Free PMC article. Review.
Cited by
-
Protective effect of nuclear factor E2-related factor 2 on inflammatory cytokine response to brominated diphenyl ether-47 in the HTR-8/SVneo human first trimester extravillous trophoblast cell line.Toxicol Appl Pharmacol. 2014 Nov 15;281(1):67-77. doi: 10.1016/j.taap.2014.09.015. Epub 2014 Oct 11. Toxicol Appl Pharmacol. 2014. PMID: 25305463 Free PMC article.
-
Non-canonical ubiquitination of the cholesterol-regulated degron of squalene monooxygenase.J Biol Chem. 2019 May 17;294(20):8134-8147. doi: 10.1074/jbc.RA119.007798. Epub 2019 Apr 2. J Biol Chem. 2019. PMID: 30940729 Free PMC article.
-
Post-translational control of the long and winding road to cholesterol.J Biol Chem. 2020 Dec 18;295(51):17549-17559. doi: 10.1074/jbc.REV120.010723. J Biol Chem. 2020. PMID: 33453997 Free PMC article. Review.
-
Twin enzymes, divergent control: The cholesterogenic enzymes DHCR14 and LBR are differentially regulated transcriptionally and post-translationally.J Biol Chem. 2020 Feb 28;295(9):2850-2865. doi: 10.1074/jbc.RA119.011323. Epub 2020 Jan 7. J Biol Chem. 2020. PMID: 31911440 Free PMC article.
-
DHCR24 associates strongly with the endoplasmic reticulum beyond predicted membrane domains: implications for the activities of this multi-functional enzyme.Biosci Rep. 2014 Apr 1;34(2):e00098. doi: 10.1042/BSR20130127. Print 2014 Apr 1. Biosci Rep. 2014. PMID: 27919032 Free PMC article.
References
-
- Waterham H. R., Koster J., Romeijn G. J., Hennekam R. C., Vreken P., Andersson H. C., FitzPatrick D. R., Kelley R. I., Wanders R. J. 2001. Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am. J. Hum. Genet. 69: 685–694 - PMC - PubMed
-
- Pedretti A., Bocci E., Maggi R., Vistoli G. 2008. Homology modelling of human DHCR24 (seladin-1) and analysis of its binding properties through molecular docking and dynamics simulations. Steroids. 73: 708–719 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

