Flow cytometry bioinformatics
- PMID: 24363631
- PMCID: PMC3867282
- DOI: 10.1371/journal.pcbi.1003365
Flow cytometry bioinformatics
Abstract
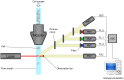
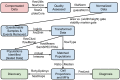
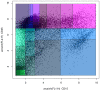
Flow cytometry bioinformatics is the application of bioinformatics to flow cytometry data, which involves storing, retrieving, organizing, and analyzing flow cytometry data using extensive computational resources and tools. Flow cytometry bioinformatics requires extensive use of and contributes to the development of techniques from computational statistics and machine learning. Flow cytometry and related methods allow the quantification of multiple independent biomarkers on large numbers of single cells. The rapid growth in the multidimensionality and throughput of flow cytometry data, particularly in the 2000s, has led to the creation of a variety of computational analysis methods, data standards, and public databases for the sharing of results. Computational methods exist to assist in the preprocessing of flow cytometry data, identifying cell populations within it, matching those cell populations across samples, and performing diagnosis and discovery using the results of previous steps. For preprocessing, this includes compensating for spectral overlap, transforming data onto scales conducive to visualization and analysis, assessing data for quality, and normalizing data across samples and experiments. For population identification, tools are available to aid traditional manual identification of populations in two-dimensional scatter plots (gating), to use dimensionality reduction to aid gating, and to find populations automatically in higher dimensional space in a variety of ways. It is also possible to characterize data in more comprehensive ways, such as the density-guided binary space partitioning technique known as probability binning, or by combinatorial gating. Finally, diagnosis using flow cytometry data can be aided by supervised learning techniques, and discovery of new cell types of biological importance by high-throughput statistical methods, as part of pipelines incorporating all of the aforementioned methods. Open standards, data, and software are also key parts of flow cytometry bioinformatics. Data standards include the widely adopted Flow Cytometry Standard (FCS) defining how data from cytometers should be stored, but also several new standards under development by the International Society for Advancement of Cytometry (ISAC) to aid in storing more detailed information about experimental design and analytical steps. Open data is slowly growing with the opening of the CytoBank database in 2010 and FlowRepository in 2012, both of which allow users to freely distribute their data, and the latter of which has been recommended as the preferred repository for MIFlowCyt-compliant data by ISAC. Open software is most widely available in the form of a suite of Bioconductor packages, but is also available for web execution on the GenePattern platform.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

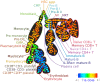


References
-
- Brando B, Barnett D, Janossy G, Mandy F, Autran B, et al. (2000) Cytofluorometric methods for assessing absolute numbers of cell subsets in blood. European Working Group on Clinical Cell Analysis. Cytometry 42: 327–346. - PubMed
-
- Ferreira-Facio CS, Milito C, Botafogo V, Fontana M, Thiago LS, et al. (2013) Contribution of multiparameter flow cytometry immunophenotyping to the diagnostic screening and classification of pediatric cancer. PLoS ONE 8: e55534 doi:10.1371/journal.pone.0055534 - DOI - PMC - PubMed
-
- Wu D, Wood BL, Fromm JR (2013) Flow cytometry for non-Hodgkin and classical Hodgkin lymphoma. Methods Mol Biol 971: 27–47. - PubMed
-
- Wang Y, Hammes F, De Roy K, Verstraete W, Boon N (2010) Past, present and future applications of flow cytometry in aquatic microbiology. Trends Biotechnol 28: 416–424. - PubMed
-
- Johnson LA, Flook JP, Look MV, Pinkel D (1987) Flow sorting of X and Y chromosome-bearing spermatozoa into two populations. Gamete Res 16: 1–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

