Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity
- PMID: 24364531
- PMCID: PMC3893830
- DOI: 10.1021/bi401042z
Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity
Abstract
Chlorite dismutases (Clds) are heme b containing oxidoreductases that convert chlorite to chloride and molecular oxygen. In order to elucidate the role of conserved heme cavity residues in the catalysis of this reaction comprehensive mutational and biochemical analyses of Cld from "Candidatus Nitrospira defluvii" (NdCld) were performed. Particularly, point mutations of the cavity-forming residues R173, K141, W145, W146, and E210 were performed. The effect of manipulation in 12 single and double mutants was probed by UV-vis spectroscopy, spectroelectrochemistry, pre-steady-state and steady-state kinetics, and X-ray crystallography. Resulting biochemical data are discussed with respect to the known crystal structure of wild-type NdCld and the variants R173A and R173K as well as the structures of R173E, W145V, W145F, and the R173Q/W146Y solved in this work. The findings allow a critical analysis of the role of these heme cavity residues in the reaction mechanism of chlorite degradation that is proposed to involve hypohalous acid as transient intermediate and formation of an O═O bond. The distal R173 is shown to be important (but not fully essential) for the reaction with chlorite, and, upon addition of cyanide, it acts as a proton acceptor in the formation of the resulting low-spin complex. The proximal H-bonding network including K141-E210-H160 keeps the enzyme in its ferric (E°' = -113 mV) and mainly five-coordinated high-spin state and is very susceptible to perturbation.
Figures


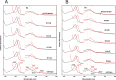

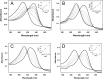

Similar articles
-
Investigation of ion binding in chlorite dismutases by means of molecular dynamics simulations.Biochemistry. 2014 Jul 29;53(29):4869-79. doi: 10.1021/bi500467h. Epub 2014 Jul 14. Biochemistry. 2014. PMID: 24988286 Free PMC article.
-
From chlorite dismutase towards HemQ - the role of the proximal H-bonding network in haeme binding.Biosci Rep. 2016 Feb 8;36(2):e00312. doi: 10.1042/BSR20150330. Biosci Rep. 2016. PMID: 26858461 Free PMC article.
-
Redox thermodynamics of high-spin and low-spin forms of chlorite dismutases with diverse subunit and oligomeric structures.Biochemistry. 2012 Nov 27;51(47):9501-12. doi: 10.1021/bi3013033. Epub 2012 Nov 14. Biochemistry. 2012. PMID: 23126649 Free PMC article.
-
Mechanism of chlorite degradation to chloride and dioxygen by the enzyme chlorite dismutase.Arch Biochem Biophys. 2015 May 15;574:18-26. doi: 10.1016/j.abb.2015.02.031. Epub 2015 Mar 4. Arch Biochem Biophys. 2015. PMID: 25748001 Review.
-
Chlorite dismutases - a heme enzyme family for use in bioremediation and generation of molecular oxygen.Biotechnol J. 2014 Apr;9(4):461-73. doi: 10.1002/biot.201300210. Epub 2014 Feb 12. Biotechnol J. 2014. PMID: 24519858 Free PMC article. Review.
Cited by
-
X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.J Biol Chem. 2020 Sep 25;295(39):13488-13501. doi: 10.1074/jbc.RA120.014087. Epub 2020 Jul 28. J Biol Chem. 2020. PMID: 32723869 Free PMC article.
-
Substrate, product, and cofactor: The extraordinarily flexible relationship between the CDE superfamily and heme.Arch Biochem Biophys. 2015 May 15;574:3-17. doi: 10.1016/j.abb.2015.03.004. Epub 2015 Mar 14. Arch Biochem Biophys. 2015. PMID: 25778630 Free PMC article. Review.
-
Structure and heme-binding properties of HemQ (chlorite dismutase-like protein) from Listeria monocytogenes.Arch Biochem Biophys. 2015 May 15;574:36-48. doi: 10.1016/j.abb.2015.01.010. Epub 2015 Jan 17. Arch Biochem Biophys. 2015. PMID: 25602700 Free PMC article.
-
Active Sites of O2-Evolving Chlorite Dismutases Probed by Halides and Hydroxides and New Iron-Ligand Vibrational Correlations.Biochemistry. 2017 Aug 29;56(34):4509-4524. doi: 10.1021/acs.biochem.7b00572. Epub 2017 Aug 17. Biochemistry. 2017. PMID: 28758386 Free PMC article.
-
Compound I Formation and Reactivity in Dimeric Chlorite Dismutase: Impact of pH and the Dynamics of the Catalytic Arginine.Biochemistry. 2023 Feb 7;62(3):835-850. doi: 10.1021/acs.biochem.2c00696. Epub 2023 Jan 27. Biochemistry. 2023. PMID: 36706455 Free PMC article.
References
-
- van Ginkel C. G.; Rikken G. B.; Kroon A. G. M.; Kengen S. W. M. (1996) Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 166, 321–326. - PubMed
-
- Maixner F.; Wagner M.; Lucker S.; Pelletier E.; Schmitz-Esser S.; Hace K.; Spieck E.; Konrat R.; Le Paslier D.; Daims H. (2008) Environmental genomics reveals functional chlorite dismutase in the nitrite-oxidizing bacterium Candidatus “Nitrospira defluvii”. Environ. Microbiol. 10, 3043–3053. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

