POMO--Plotting Omics analysis results for Multiple Organisms
- PMID: 24365393
- PMCID: PMC3880012
- DOI: 10.1186/1471-2164-14-918
POMO--Plotting Omics analysis results for Multiple Organisms
Abstract
Background: Systems biology experiments studying different topics and organisms produce thousands of data values across different types of genomic data. Further, data mining analyses are yielding ranked and heterogeneous results and association networks distributed over the entire genome. The visualization of these results is often difficult and standalone web tools allowing for custom inputs and dynamic filtering are limited.
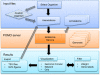
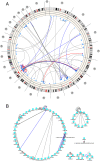
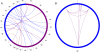
Results: We have developed POMO (http://pomo.cs.tut.fi), an interactive web-based application to visually explore omics data analysis results and associations in circular, network and grid views. The circular graph represents the chromosome lengths as perimeter segments, as a reference outer ring, such as cytoband for human. The inner arcs between nodes represent the uploaded network. Further, multiple annotation rings, for example depiction of gene copy number changes, can be uploaded as text files and represented as bar, histogram or heatmap rings. POMO has built-in references for human, mouse, nematode, fly, yeast, zebrafish, rice, tomato, Arabidopsis, and Escherichia coli. In addition, POMO provides custom options that allow integrated plotting of unsupported strains or closely related species associations, such as human and mouse orthologs or two yeast wild types, studied together within a single analysis. The web application also supports interactive label and weight filtering. Every iterative filtered result in POMO can be exported as image file and text file for sharing or direct future input.
Conclusions: The POMO web application is a unique tool for omics data analysis, which can be used to visualize and filter the genome-wide networks in the context of chromosomal locations as well as multiple network layouts. With the several illustration and filtering options the tool supports the analysis and visualization of any heterogeneous omics data analysis association results for many organisms. POMO is freely available and does not require any installation or registration.
Figures

References
-
- Palsson B, Zengler K. The challenges of integrating multi-omic data sets. Nat Chem Biol. 2010;14:787–789. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

