Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1
- PMID: 24366543
- PMCID: PMC4023822
- DOI: 10.1128/MCB.01191-13
Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1
Abstract
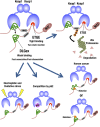
Transcription factor Nrf2 (NF-E2-related factor 2) coordinately regulates cytoprotective gene expression, but under unstressed conditions, Nrf2 is degraded rapidly through Keap1 (Kelch-like ECH-associated protein 1)-mediated ubiquitination. Nrf2 harbors two Keap1-binding motifs, DLG and ETGE. Interactions between these two motifs and Keap1 constitute a key regulatory nexus for cellular Nrf2 activity through the formation of a two-site binding hinge-and-latch mechanism. In this study, we determined the minimum Keap1-binding sequence of the DLG motif, the low-affinity latch site, and defined a new DLGex motif that covers a sequence much longer than that previously defined. We have successfully clarified the crystal structure of the Keap1-DC-DLGex complex at 1.6 Å. DLGex possesses a complicated helix structure, which interprets well the human-cancer-derived loss-of-function mutations in DLGex. In thermodynamic analyses, Keap1-DLGex binding is characterized as enthalpy and entropy driven, while Keap1-ETGE binding is characterized as purely enthalpy driven. In kinetic analyses, Keap1-DLGex binding follows a fast-association and fast-dissociation model, while Keap1-ETGE binding contains a slow-reaction step that leads to a stable conformation. These results demonstrate that the mode of DLGex binding to Keap1 is distinct from that of ETGE structurally, thermodynamically, and kinetically and support our contention that the DLGex motif serves as a converter transmitting environmental stress to Nrf2 induction as the latch site.
Figures





Similar articles
-
Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response.Mol Cell Biol. 2007 Nov;27(21):7511-21. doi: 10.1128/MCB.00753-07. Epub 2007 Sep 4. Mol Cell Biol. 2007. PMID: 17785452 Free PMC article.
-
Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model.Mol Cell Biol. 2006 Apr;26(8):2887-900. doi: 10.1128/MCB.26.8.2887-2900.2006. Mol Cell Biol. 2006. PMID: 16581765 Free PMC article.
-
Molecular basis for the disruption of Keap1-Nrf2 interaction via Hinge & Latch mechanism.Commun Biol. 2021 May 14;4(1):576. doi: 10.1038/s42003-021-02100-6. Commun Biol. 2021. PMID: 33990683 Free PMC article.
-
Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome.Arch Biochem Biophys. 2005 Jan 15;433(2):342-50. doi: 10.1016/j.abb.2004.10.012. Arch Biochem Biophys. 2005. PMID: 15581590 Review.
-
Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution.Genes Cells. 2011 Feb;16(2):123-40. doi: 10.1111/j.1365-2443.2010.01473.x. Genes Cells. 2011. PMID: 21251164 Review.
Cited by
-
Cadmium and Secondary Structure-dependent Function of a Degron in the Pca1p Cadmium Exporter.J Biol Chem. 2016 Jun 3;291(23):12420-31. doi: 10.1074/jbc.M116.724930. Epub 2016 Apr 8. J Biol Chem. 2016. PMID: 27059957 Free PMC article.
-
Nrf2 Pathway and Autophagy Crosstalk: New Insights into Therapeutic Strategies for Ischemic Cerebral Vascular Diseases.Antioxidants (Basel). 2022 Sep 2;11(9):1747. doi: 10.3390/antiox11091747. Antioxidants (Basel). 2022. PMID: 36139821 Free PMC article. Review.
-
Investigation of Molecular Details of Keap1-Nrf2 Inhibitors Using Molecular Dynamics and Umbrella Sampling Techniques.Molecules. 2019 Nov 12;24(22):4085. doi: 10.3390/molecules24224085. Molecules. 2019. PMID: 31726716 Free PMC article.
-
Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):E12218-E12227. doi: 10.1073/pnas.1818012115. Epub 2018 Dec 11. Proc Natl Acad Sci U S A. 2018. PMID: 30538201 Free PMC article.
-
The protein kinase activity of fructokinase A specifies the antioxidant responses of tumor cells by phosphorylating p62.Sci Adv. 2019 Apr 24;5(4):eaav4570. doi: 10.1126/sciadv.aav4570. eCollection 2019 Apr. Sci Adv. 2019. PMID: 31032410 Free PMC article.
References
-
- Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. 2006. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol. Chem. 387:1311–1320 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
