Increased diversity of the HLA-B40 ligandome by the presentation of peptides phosphorylated at their main anchor residue
- PMID: 24366607
- PMCID: PMC3916647
- DOI: 10.1074/mcp.M113.034314
Increased diversity of the HLA-B40 ligandome by the presentation of peptides phosphorylated at their main anchor residue
Abstract
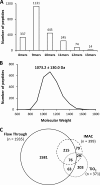
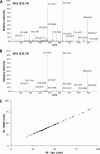
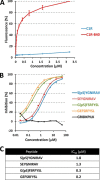
Human leukocyte antigen (HLA) class I molecules bind peptides derived from the intracellular degradation of endogenous proteins and present them to cytotoxic T lymphocytes, allowing the immune system to detect transformed or virally infected cells. It is known that HLA class I-associated peptides may harbor posttranslational modifications. In particular, phosphorylated ligands have raised much interest as potential targets for cancer immunotherapy. By combining affinity purification with high-resolution mass spectrometry, we identified more than 2000 unique ligands bound to HLA-B40. Sequence analysis revealed two major anchor motifs: aspartic or glutamic acid at peptide position 2 (P2) and methionine, phenylalanine, or aliphatic residues at the C terminus. The use of immobilized metal ion and TiO2 affinity chromatography allowed the characterization of 85 phosphorylated ligands. We further confirmed every sequence belonging to this subset by comparing its experimental MS2 spectrum with that obtained upon fragmentation of the corresponding synthetic peptide. Remarkably, three phospholigands lacked a canonical anchor residue at P2, containing phosphoserine instead. Binding assays showed that these peptides bound to HLA-B40 with high affinity. Together, our data demonstrate that the peptidome of a given HLA allotype can be broadened by the presentation of peptides with posttranslational modifications at major anchor positions. We suggest that ligands with phosphorylated residues at P2 might be optimal targets for T-cell-based cancer immunotherapy.
Figures



Similar articles
-
A Molecular Basis for the Presentation of Phosphorylated Peptides by HLA-B Antigens.Mol Cell Proteomics. 2017 Feb;16(2):181-193. doi: 10.1074/mcp.M116.063800. Epub 2016 Dec 5. Mol Cell Proteomics. 2017. PMID: 27920218 Free PMC article.
-
Comparative Analysis of the Endogenous Peptidomes Displayed by HLA-B*27 and Mamu-B*08: Two MHC Class I Alleles Associated with Elite Control of HIV/SIV Infection.J Proteome Res. 2016 Mar 4;15(3):1059-69. doi: 10.1021/acs.jproteome.5b01146. Epub 2016 Feb 8. J Proteome Res. 2016. PMID: 26811146
-
Ligandomes obtained from different HLA-class II-molecules are homologous for N- and C-terminal residues outside the peptide-binding cleft.Immunogenetics. 2019 Sep;71(8-9):519-530. doi: 10.1007/s00251-019-01129-6. Epub 2019 Sep 13. Immunogenetics. 2019. PMID: 31520135 Free PMC article.
-
What Is the Role of HLA-I on Cancer Derived Extracellular Vesicles? Defining the Challenges in Characterisation and Potential Uses of This Ligandome.Int J Mol Sci. 2021 Dec 17;22(24):13554. doi: 10.3390/ijms222413554. Int J Mol Sci. 2021. PMID: 34948350 Free PMC article. Review.
-
Soluble HLA peptidome: A new resource for cancer biomarkers.Front Oncol. 2022 Dec 22;12:1069635. doi: 10.3389/fonc.2022.1069635. eCollection 2022. Front Oncol. 2022. PMID: 36620582 Free PMC article. Review.
Cited by
-
An open-source computational and data resource to analyze digital maps of immunopeptidomes.Elife. 2015 Jul 8;4:e07661. doi: 10.7554/eLife.07661. Elife. 2015. PMID: 26154972 Free PMC article.
-
Recent advances in enrichment and separation strategies for mass spectrometry-based phosphoproteomics.Electrophoresis. 2014 Dec;35(24):3418-29. doi: 10.1002/elps.201400017. Epub 2014 Jun 16. Electrophoresis. 2014. PMID: 24687451 Free PMC article. Review.
-
The Evolving Landscape of Autoantigen Discovery and Characterization in Type 1 Diabetes.Diabetes. 2019 May;68(5):879-886. doi: 10.2337/dbi18-0066. Diabetes. 2019. PMID: 31010879 Free PMC article.
-
Substantial Influence of ERAP2 on the HLA-B*40:02 Peptidome: Implications for HLA-B*27-Negative Ankylosing Spondylitis.Mol Cell Proteomics. 2019 Nov;18(11):2298-2309. doi: 10.1074/mcp.RA119.001710. Epub 2019 Sep 17. Mol Cell Proteomics. 2019. PMID: 31530632 Free PMC article.
-
Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes Using Mass Spectrometry.Mol Cell Proteomics. 2015 Dec;14(12):3105-17. doi: 10.1074/mcp.O115.052431. Mol Cell Proteomics. 2015. PMID: 26628741 Free PMC article. Review.
References
-
- Madden D. R. (1995) The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13, 587–622 - PubMed
-
- Garrett T. P., Saper M. A., Bjorkman P. J., Strominger J. L., Wiley D. C. (1989) Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature 342, 692–696 - PubMed
-
- Brusic V., Bajic V. B., Petrovsky N. (2004) Computational methods for prediction of T-cell epitopes—a framework for modelling, testing, and applications. Methods 34, 436–443 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

