Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae
- PMID: 24367018
- PMCID: PMC3912082
- DOI: 10.1104/pp.113.227637
Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae
Abstract
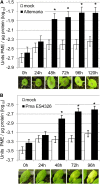
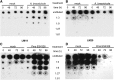
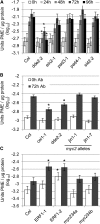
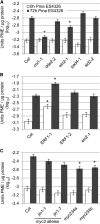
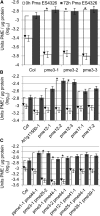
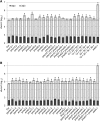
Pectins, major components of dicot cell walls, are synthesized in a heavily methylesterified form in the Golgi and are partially deesterified by pectin methylesterases (PMEs) upon export to the cell wall. PME activity is important for the virulence of the necrotrophic fungal pathogen Botrytis cinerea. Here, the roles of Arabidopsis PMEs in pattern-triggered immunity and immune responses to the necrotrophic fungus Alternaria brassicicola and the bacterial hemibiotroph Pseudomonas syringae pv maculicola ES4326 (Pma ES4326) were studied. Plant PME activity increased during pattern-triggered immunity and after inoculation with either pathogen. The increase of PME activity in response to pathogen treatment was concomitant with a decrease in pectin methylesterification. The pathogen-induced PME activity did not require salicylic acid or ethylene signaling, but was dependent on jasmonic acid signaling. In the case of induction by A. brassicicola, the ethylene response factor, but not the MYC2 branch of jasmonic acid signaling, contributed to induction of PME activity, whereas in the case of induction by Pma ES4326, both branches contributed. There are 66 PME genes in Arabidopsis, suggesting extensive genetic redundancy. Nevertheless, selected pme single, double, triple and quadruple mutants allowed significantly more growth of Pma ES4326 than wild-type plants, indicating a role of PMEs in resistance to this pathogen. No decreases in total PME activity were detected in these pme mutants, suggesting that the determinant of immunity is not total PME activity; rather, it is some specific effect of PMEs such as changes in the pattern of pectin methylesterification.
Figures
References
-
- Albersheim P, Jones TM, English PD. (1969) Biochemistry of the cell wall in relation to infective processes. Annu Rev Phytopathol 7: 171–194 - PubMed
-
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

