Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces
- PMID: 24367152
- PMCID: PMC3859367
- DOI: 10.2183/pjab/83.277
Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces
Abstract
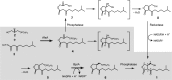
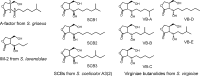
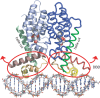
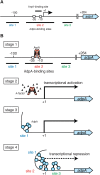
Streptomyces griseus, a well-known industrial producer of streptomycin, is a member of the genus Streptomyces, which shows a complex life cycle resembling that of fungi. A-factor, a C13 γ-butyrolactone compound, was discovered as a self-regulatory factor or a bacterial hormone to induce morphological differentiation and production of secondary metabolites, including streptomycin, in this organism. Accumulating evidence has revealed an A-factor-triggered signal cascade, which is composed of several key steps or components. These include: (i) AfsA catalyzing a crucial step of A-factor biosynthesis, (ii) the A-factor-specific receptor (ArpA), which acts as a transcriptional repressor for adpA, (iii) adpA, a sole target of ArpA, which encodes a global transcriptional activator AdpA, and (iv) a variety of members of the AdpA regulon, a set of the genes regulated by AdpA. A-factor is biosynthesized via five reaction steps, in which AfsA catalyzes acyl transfer between a β-ketoacyl-acyl carrier protein and the hydroxyl group of dihydroxyacetone phosphate. The receptor ArpA, belonging to the TetR family, is a homodimer, each subunit of which contains a helix-turn-helix DNA-binding motif and an A-factor-binding pocket. The three-dimensional structure and conformational change upon binding A-factor are elucidated, on the basis of X-ray crystallography of CprB, an ArpA homologue. AdpA, belonging to the AraC/XylS transcriptional activator family, binds operators upstream from the promoters of a variety of the target genes and activates their transcription, thus forming the AdpA regulon. Members of the AdpA regulon includes the pathway-specific transcriptional activator gene strR that activates the whole streptomycin biosynthesis gene cluster, in addition to a number of genes that direct the multiple cellular functions required for cellular differentiation in a concerted manner. A variety of A-factor homologues as well as homologues of afsA/arpA are distributed widely among Streptomyces, indicating the significant role of this type of molecular signaling in the ecosystem and evolutional processes.
Keywords: A-factor; A-factor receptor; Streptomyces; morphological differentiation; secondary metabolism; streptomycin biosynthesis.
Figures






References
-
- Hopwood, D. A. (1999) Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145, 2183–2202 - PubMed
-
- Horinouchi, S. and Beppu, T. (1994) A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12, 859–864 - PubMed
-
- Ohnishi, Y., Yamazaki, H., Kato, J., Tomono, A. and Horinouchi, S. (2005) AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69, 431–439 - PubMed
-
- Horinouchi, S. (2007) Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci. Biotechnol. Biochem. 71, 283–299 - PubMed
-
- Khokhlov, A. S., Tovarova, I. I., Borisova, L. N., Pliner, S. A., Schevchenko, L. A., Kornitskaya, E. Y., Ivkina, N. S. and Rapoport, I. A. (1967) A-factor responsible for the biosynthesis of streptomycin by a mutant strain of Actinomyces streptomycini. Dokl. Akad. Nauk SSSR 177, 232–235 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
