Transcriptional analysis of murine macrophages infected with different Toxoplasma strains identifies novel regulation of host signaling pathways
- PMID: 24367253
- PMCID: PMC3868521
- DOI: 10.1371/journal.ppat.1003779
Transcriptional analysis of murine macrophages infected with different Toxoplasma strains identifies novel regulation of host signaling pathways
Abstract
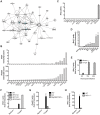
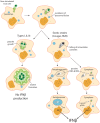
Most isolates of Toxoplasma from Europe and North America fall into one of three genetically distinct clonal lineages, the type I, II and III lineages. However, in South America these strains are rarely isolated and instead a great variety of other strains are found. T. gondii strains differ widely in a number of phenotypes in mice, such as virulence, persistence, oral infectivity, migratory capacity, induction of cytokine expression and modulation of host gene expression. The outcome of toxoplasmosis in patients is also variable and we hypothesize that, besides host and environmental factors, the genotype of the parasite strain plays a major role. The molecular basis for these differences in pathogenesis, especially in strains other than the clonal lineages, remains largely unexplored. Macrophages play an essential role in the early immune response against T. gondii and are also the cell type preferentially infected in vivo. To determine if non-canonical Toxoplasma strains have unique interactions with the host cell, we infected murine macrophages with 29 different Toxoplasma strains, representing global diversity, and used RNA-sequencing to determine host and parasite transcriptomes. We identified large differences between strains in the expression level of known parasite effectors and large chromosomal structural variation in some strains. We also identified novel strain-specifically regulated host pathways, including the regulation of the type I interferon response by some atypical strains. IFNβ production by infected cells was associated with parasite killing, independent of interferon gamma activation, and dependent on endosomal Toll-like receptors in macrophages and the cytoplasmic receptor retinoic acid-inducible gene 1 (RIG-I) in fibroblasts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

