HrpA, an RNA helicase involved in RNA processing, is required for mouse infectivity and tick transmission of the Lyme disease spirochete
- PMID: 24367266
- PMCID: PMC3868530
- DOI: 10.1371/journal.ppat.1003841
HrpA, an RNA helicase involved in RNA processing, is required for mouse infectivity and tick transmission of the Lyme disease spirochete
Abstract
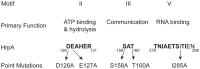
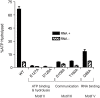
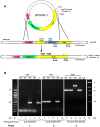
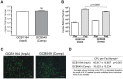
The Lyme disease spirochete Borrelia burgdorferi must differentially express genes and proteins in order to survive in and transit between its tick vector and vertebrate reservoir. The putative DEAH-box RNA helicase, HrpA, has been recently identified as an addition to the spirochete's global regulatory machinery; using proteomic methods, we demonstrated that HrpA modulates the expression of at least 180 proteins. Although most bacteria encode an HrpA helicase, RNA helicase activity has never been demonstrated for HrpAs and the literature contains little information on the contribution of this protein to bacterial physiology or pathogenicity. In this work, we report that B. burgdorferi HrpA has RNA-stimulated ATPase activity and RNA helicase activity and that this enzyme is essential for both mammalian infectivity by syringe inoculation and tick transmission. Reduced infectivity of strains carrying mutations in the ATPase and RNA binding motif mutants suggests that full virulence expression requires both ATPase and coupled helicase activity. Microarray profiling revealed changes in RNA levels of two-fold, or less in an hrpA mutant versus wild-type, suggesting that the enzyme functions largely or exclusively at the post-transcriptional level. In this regard, northern blot analysis of selected gene products highly regulated by HrpA (bb0603 [p66], bba74, bb0241 [glpK], bb0242 and bb0243 [glpA]) suggests a role for HrpA in the processing and translation of transcripts. In addition to being the first demonstration of RNA helicase activity for a bacterial HrpA, our data indicate that the post-transcriptional regulatory functions of this enzyme are essential for maintenance of the Lyme disease spirochete's enzootic cycle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
HrpA, a DEAH-box RNA helicase, is involved in global gene regulation in the Lyme disease spirochete.PLoS One. 2011;6(7):e22168. doi: 10.1371/journal.pone.0022168. Epub 2011 Jul 26. PLoS One. 2011. PMID: 21814569 Free PMC article.
-
The Infectivity Gene bbk13 Is Important for Multiple Phases of the Borrelia burgdorferi Enzootic Cycle.Infect Immun. 2021 Sep 16;89(10):e0021621. doi: 10.1128/IAI.00216-21. Epub 2021 Jun 28. Infect Immun. 2021. PMID: 34181460 Free PMC article.
-
Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation.Mol Microbiol. 2015 Feb;95(3):509-38. doi: 10.1111/mmi.12882. Epub 2014 Dec 30. Mol Microbiol. 2015. PMID: 25425211 Free PMC article.
-
Borrelia burgdorferi--traveling incognito?Microbes Infect. 2006 Apr;8(5):1390-9. doi: 10.1016/j.micinf.2005.12.022. Epub 2006 Mar 24. Microbes Infect. 2006. PMID: 16698304 Review.
-
Insights into the biology of Borrelia burgdorferi gained through the application of molecular genetics.Adv Appl Microbiol. 2014;86:41-143. doi: 10.1016/B978-0-12-800262-9.00002-0. Adv Appl Microbiol. 2014. PMID: 24377854 Review.
Cited by
-
Extragenic Suppression of Elongation Factor P Gene Mutant Phenotypes in Erwinia amylovora.J Bacteriol. 2019 May 8;201(11):e00722-18. doi: 10.1128/JB.00722-18. Print 2019 Jun 1. J Bacteriol. 2019. PMID: 30885930 Free PMC article.
-
Borrelia burgdorferi HtrA: evidence for twofold proteolysis of outer membrane protein p66.Mol Microbiol. 2016 Jan;99(1):135-50. doi: 10.1111/mmi.13221. Epub 2015 Oct 20. Mol Microbiol. 2016. PMID: 26370492 Free PMC article.
-
Lyme Disease Pathogenesis.Curr Issues Mol Biol. 2021;42:473-518. doi: 10.21775/cimb.042.473. Epub 2020 Dec 23. Curr Issues Mol Biol. 2021. PMID: 33353871 Free PMC article. Review.
-
DExD-box RNA-helicases in Listeria monocytogenes are important for growth, ribosomal maturation, rRNA processing and virulence factor expression.RNA Biol. 2014;11(11):1457-66. doi: 10.1080/15476286.2014.996099. RNA Biol. 2014. PMID: 25590644 Free PMC article.
-
PA3297 Counteracts Antimicrobial Effects of Azithromycin in Pseudomonas aeruginosa.Front Microbiol. 2016 Mar 16;7:317. doi: 10.3389/fmicb.2016.00317. eCollection 2016. Front Microbiol. 2016. PMID: 27014238 Free PMC article.
References
-
- Stanek G, Wormser GP, Gray J, Strle F (2012) Lyme borreliosis. Lancet 379: 461–473. - PubMed
-
- Skare JT, Carroll JA, X.F Y, Samuels DS, Akins DR (2010) Gene regulation, transcriptomics and proteomics. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Norfolk, UK: Caister Academic Press. pp. 67–101.
-
- Samuels DS (2011) Gene Regulation in Borrelia burgdorferi . Annu Rev Microbiol - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

