Base pairing interaction between 5'- and 3'-UTRs controls icaR mRNA translation in Staphylococcus aureus
- PMID: 24367275
- PMCID: PMC3868564
- DOI: 10.1371/journal.pgen.1004001
Base pairing interaction between 5'- and 3'-UTRs controls icaR mRNA translation in Staphylococcus aureus
Abstract
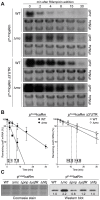
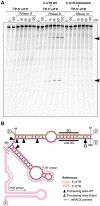
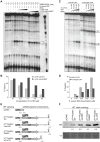
The presence of regulatory sequences in the 3' untranslated region (3'-UTR) of eukaryotic mRNAs controlling RNA stability and translation efficiency is widely recognized. In contrast, the relevance of 3'-UTRs in bacterial mRNA functionality has been disregarded. Here, we report evidences showing that around one-third of the mapped mRNAs of the major human pathogen Staphylococcus aureus carry 3'-UTRs longer than 100-nt and thus, potential regulatory functions. We selected the long 3'-UTR of icaR, which codes for the repressor of the main exopolysaccharidic compound of the S. aureus biofilm matrix, to evaluate the role that 3'-UTRs may play in controlling mRNA expression. We showed that base pairing between the 3'-UTR and the Shine-Dalgarno (SD) region of icaR mRNA interferes with the translation initiation complex and generates a double-stranded substrate for RNase III. Deletion or substitution of the motif (UCCCCUG) within icaR 3'-UTR was sufficient to abolish this interaction and resulted in the accumulation of IcaR repressor and inhibition of biofilm development. Our findings provide a singular example of a new potential post-transcriptional regulatory mechanism to modulate bacterial gene expression through the interaction of a 3'-UTR with the 5'-UTR of the same mRNA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Regulation of Tacaribe Mammarenavirus Translation: Positive 5' and Negative 3' Elements and Role of Key Cellular Factors.J Virol. 2017 Jun 26;91(14):e00084-17. doi: 10.1128/JVI.00084-17. Print 2017 Jul 15. J Virol. 2017. PMID: 28468879 Free PMC article.
-
Translation enhancer in the 3'-untranslated region of rotavirus gene 6 mRNA promotes expression of the major capsid protein VP6.Arch Virol. 2004 Feb;149(2):303-21. doi: 10.1007/s00705-003-0211-9. Epub 2003 Oct 30. Arch Virol. 2004. PMID: 14745597
-
The 5'-untranslated region of the ntp303 gene strongly enhances translation during pollen tube growth, but not during pollen maturation.Plant Physiol. 2002 May;129(1):342-53. doi: 10.1104/pp.001701. Plant Physiol. 2002. PMID: 12011364 Free PMC article.
-
The Untranslated Regions of mRNAs in Cancer.Trends Cancer. 2019 Apr;5(4):245-262. doi: 10.1016/j.trecan.2019.02.011. Epub 2019 Mar 22. Trends Cancer. 2019. PMID: 30961831 Free PMC article. Review.
-
3'-Untranslated regions are important in mRNA localization and translation: lessons from selenium and metallothionein.Biochem Soc Trans. 2004 Dec;32(Pt 6):990-3. doi: 10.1042/BST0320990. Biochem Soc Trans. 2004. PMID: 15506944 Review.
Cited by
-
Plasmid-Chromosome Crosstalk in Staphylococcus aureus: A Horizontally Acquired Transcription Regulator Controls Polysaccharide Intercellular Adhesin-Mediated Biofilm Formation.Front Cell Infect Microbiol. 2021 Mar 22;11:660702. doi: 10.3389/fcimb.2021.660702. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33829001 Free PMC article.
-
A eukaryotic-like 3' untranslated region in Salmonella enterica hilD mRNA.Nucleic Acids Res. 2014 May;42(9):5894-906. doi: 10.1093/nar/gku222. Epub 2014 Mar 20. Nucleic Acids Res. 2014. PMID: 24682814 Free PMC article.
-
Complex Regulation of Gamma-Hemolysin Expression Impacts Staphylococcus aureus Virulence.Microbiol Spectr. 2023 Aug 17;11(4):e0107323. doi: 10.1128/spectrum.01073-23. Epub 2023 Jun 22. Microbiol Spectr. 2023. PMID: 37347186 Free PMC article.
-
A high-resolution view of RNA endonuclease cleavage in Bacillus subtilis.Nucleic Acids Res. 2025 Jan 24;53(3):gkaf030. doi: 10.1093/nar/gkaf030. Nucleic Acids Res. 2025. PMID: 39883015 Free PMC article.
-
Widespread formation of alternative 3' UTR isoforms via transcription termination in archaea.Nat Microbiol. 2016 Aug 22;1(10):16143. doi: 10.1038/nmicrobiol.2016.143. Nat Microbiol. 2016. PMID: 27670118
References
-
- Mazumder B, Seshadri V, Fox PL (2003) Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci 28: 91–98 doi:10.1016/S0968-0004(03)00002-1 - DOI - PubMed
-
- Brodersen P, Voinnet O (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10: 141–148 doi:10.1038/nrm2619 - DOI - PubMed
-
- Mangus DA, Evans MC, Jacobson A (2003) Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223 doi:10.1186/gb-2003-4-7-223 - DOI - PMC - PubMed
-
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 doi:10.1038/nrm2104 - DOI - PubMed
-
- Matoulkova E, Michalova E, Vojtesek B, Hrstka R (2012) The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol 9: 563–576 doi:10.4161/rna.20231 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

