Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation
- PMID: 24367281
- PMCID: PMC3868596
- DOI: 10.1371/journal.pgen.1004033
Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation
Abstract
More than 95% of the human population is infected with human herpesvirus-6 (HHV-6) during early childhood and maintains latent HHV-6 genomes either in an extra-chromosomal form or as a chromosomally integrated HHV-6 (ciHHV-6). In addition, approximately 1% of humans are born with an inheritable form of ciHHV-6 integrated into the telomeres of chromosomes. Immunosuppression and stress conditions can reactivate latent HHV-6 replication, which is associated with clinical complications and even death. We have previously shown that Chlamydia trachomatis infection reactivates ciHHV-6 and induces the formation of extra-chromosomal viral DNA in ciHHV-6 cells. Here, we propose a model and provide experimental evidence for the mechanism of ciHHV-6 reactivation. Infection with Chlamydia induced a transient shortening of telomeric ends, which subsequently led to increased telomeric circle (t-circle) formation and incomplete reconstitution of circular viral genomes containing single viral direct repeat (DR). Correspondingly, short t-circles containing parts of the HHV-6 DR were detected in cells from individuals with genetically inherited ciHHV-6. Furthermore, telomere shortening induced in the absence of Chlamydia infection also caused circularization of ciHHV-6, supporting a t-circle based mechanism for ciHHV-6 reactivation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
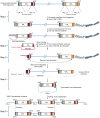
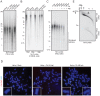

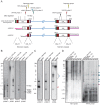
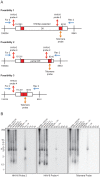
References
-
- Tanaka-Taya K, Sashihara J, Kurahashi H, Amo K, Miyagawa H, et al. (2004) Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol 73: 465–473. - PubMed
-
- Daibata M, Taguchi T, Nemoto Y, Taguchi H, Miyoshi I (1999) Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 94: 1545. - PubMed
-
- Clark DA (2000) Human herpesvirus 6. Rev Med Virol 10: 155–173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

