Structural context of disease-associated mutations and putative mechanism of autoinhibition revealed by X-ray crystallographic analysis of the EZH2-SET domain
- PMID: 24367637
- PMCID: PMC3868555
- DOI: 10.1371/journal.pone.0084147
Structural context of disease-associated mutations and putative mechanism of autoinhibition revealed by X-ray crystallographic analysis of the EZH2-SET domain
Abstract
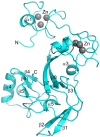
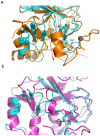
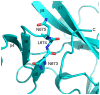
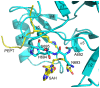
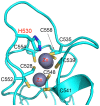
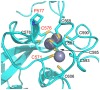
The enhancer-of-zeste homolog 2 (EZH2) gene product is an 87 kDa polycomb group (PcG) protein containing a C-terminal methyltransferase SET domain. EZH2, along with binding partners, i.e., EED and SUZ12, upon which it is dependent for activity forms the core of the polycomb repressive complex 2 (PRC2). PRC2 regulates gene silencing by catalyzing the methylation of histone H3 at lysine 27. Both overexpression and mutation of EZH2 are associated with the incidence and aggressiveness of various cancers. The novel crystal structure of the SET domain was determined in order to understand disease-associated EZH2 mutations and derive an explanation for its inactivity independent of complex formation. The 2.00 Å crystal structure reveals that, in its uncomplexed form, the EZH2 C-terminus folds back into the active site blocking engagement with substrate. Furthermore, the S-adenosyl-L-methionine (SAM) binding pocket observed in the crystal structure of homologous SET domains is notably absent. This suggests that a conformational change in the EZH2 SET domain, dependent upon complex formation, must take place for cofactor and substrate binding activities to be recapitulated. In addition, the data provide a structural context for clinically significant mutations found in the EZH2 SET domain.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

