Genome-wide analyses of radioresistance-associated miRNA expression profile in nasopharyngeal carcinoma using next generation deep sequencing
- PMID: 24367666
- PMCID: PMC3868612
- DOI: 10.1371/journal.pone.0084486
Genome-wide analyses of radioresistance-associated miRNA expression profile in nasopharyngeal carcinoma using next generation deep sequencing
Erratum in
- PLoS One. 2014;9(10):e110099
Abstract
Background: Rapidly growing evidence suggests that microRNAs (miRNAs) are involved in a wide range of cancer malignant behaviours including radioresistance. Therefore, the present study was designed to investigate miRNA expression patterns associated with radioresistance in NPC.
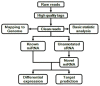
Methods: The differential expression profiles of miRNAs and mRNAs associated with NPC radioresistance were constructed. The predicted target mRNAs of miRNAs and their enriched signaling pathways were analyzed via biological informatical algorithms. Finally, partial miRNAs and pathways-correlated target mRNAs were validated in two NPC radioreisitant cell models.
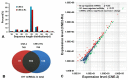
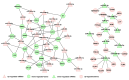
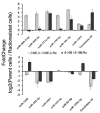
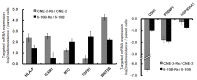
Results: 50 known and 9 novel miRNAs with significant difference were identified, and their target mRNAs were narrowed down to 53 nasopharyngeal-/NPC-specific mRNAs. Subsequent KEGG analyses demonstrated that the 53 mRNAs were enriched in 37 signaling pathways. Further qRT-PCR assays confirmed 3 down-regulated miRNAs (miR-324-3p, miR-93-3p and miR-4501), 3 up-regulated miRNAs (miR-371a-5p, miR-34c-5p and miR-1323) and 2 novel miRNAs. Additionally, corresponding alterations of pathways-correlated target mRNAs were observed including 5 up-regulated mRNAs (ICAM1, WNT2B, MYC, HLA-F and TGF-β1) and 3 down-regulated mRNAs (CDH1, PTENP1 and HSP90AA1).
Conclusions: Our study provides an overview of miRNA expression profile and the interactions between miRNA and their target mRNAs, which will deepen our understanding of the important roles of miRNAs in NPC radioresistance.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

