Regulated aggregative multicellularity in a close unicellular relative of metazoa
- PMID: 24368732
- PMCID: PMC3870316
- DOI: 10.7554/eLife.01287
Regulated aggregative multicellularity in a close unicellular relative of metazoa
Abstract
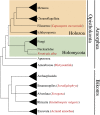
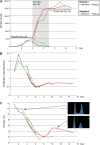
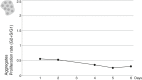
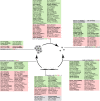
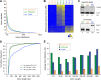
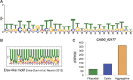
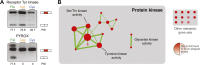
The evolution of metazoans from their unicellular ancestors was one of the most important events in the history of life. However, the cellular and genetic changes that ultimately led to the evolution of multicellularity are not known. In this study, we describe an aggregative multicellular stage in the protist Capsaspora owczarzaki, a close unicellular relative of metazoans. Remarkably, transition to the aggregative stage is associated with significant upregulation of orthologs of genes known to establish multicellularity and tissue architecture in metazoans. We further observe transitions in regulated alternative splicing during the C. owczarzaki life cycle, including the deployment of an exon network associated with signaling, a feature of splicing regulation so far only observed in metazoans. Our results reveal the existence of a highly regulated aggregative stage in C. owczarzaki and further suggest that features of aggregative behavior in an ancestral protist may had been co-opted to develop some multicellular properties currently seen in metazoans. DOI: http://dx.doi.org/10.7554/eLife.01287.001.
Keywords: Capsaspora; alternative splicing; cell differentiation; evolutionary transitions; opisthokonts.
Conflict of interest statement
BJB: Reviewing editor,
The other authors declare that no competing interests exist.
Figures







Comment in
-
Multicellularity: From brief encounters to lifelong unions.Elife. 2013 Dec 24;2:e01893. doi: 10.7554/eLife.01893. Elife. 2013. PMID: 24368736 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

