Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis
- PMID: 24368785
- PMCID: PMC3904003
- DOI: 10.1105/tpc.113.120162
Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis
Abstract
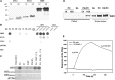
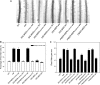
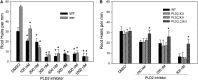
Phosphatidic acid (PA) has emerged as a class of cellular mediators involved in various cellular and physiological processes, but little is known about its mechanism of action. Here we show that PA interacts with werewolf (WER), a R2R3 MYB transcription factor involved in root hair formation. The PA-interacting region is confined to the end of the R2 subdomain. The ablation of the PA binding motif has no effect on WER binding to DNA, but abolishes its nuclear localization and its function in regulating epidermal cell fate. Inhibition of PA production by phospholipase Dζ also suppresses WER's nuclear localization, root hair formation, and elongation. These results suggest a role for PA in promoting protein nuclear localization.
Figures





Similar articles
-
Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis.Plant Cell. 2007 Jul;19(7):2264-77. doi: 10.1105/tpc.106.045732. Epub 2007 Jul 20. Plant Cell. 2007. PMID: 17644729 Free PMC article.
-
The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis.Development. 2005 Nov;132(21):4765-75. doi: 10.1242/dev.02055. Epub 2005 Oct 5. Development. 2005. PMID: 16207757
-
Structural insights into target DNA recognition by R2R3-MYB transcription factors.Nucleic Acids Res. 2020 Jan 10;48(1):460-471. doi: 10.1093/nar/gkz1081. Nucleic Acids Res. 2020. PMID: 31733060 Free PMC article.
-
[The mechanism of root hair development and molecular regulation in plants].Yi Chuan. 2007 Apr;29(4):413-9. doi: 10.1360/yc-007-0413. Yi Chuan. 2007. PMID: 17548302 Review. Chinese.
-
GLABRA2, A Common Regulator for Epidermal Cell Fate Determination and Anthocyanin Biosynthesis in Arabidopsis.Int J Mol Sci. 2019 Oct 9;20(20):4997. doi: 10.3390/ijms20204997. Int J Mol Sci. 2019. PMID: 31601032 Free PMC article. Review.
Cited by
-
Dynamic responses of PA to environmental stimuli imaged by a genetically encoded mobilizable fluorescent sensor.Plant Commun. 2023 May 8;4(3):100500. doi: 10.1016/j.xplc.2022.100500. Epub 2022 Nov 29. Plant Commun. 2023. PMID: 36447433 Free PMC article.
-
Phospholipase pPLAIIIα Increases Germination Rate and Resistance to Turnip Crinkle Virus when Overexpressed.Plant Physiol. 2020 Nov;184(3):1482-1498. doi: 10.1104/pp.20.00630. Epub 2020 Aug 28. Plant Physiol. 2020. PMID: 32859754 Free PMC article.
-
Phloem Proteomics Reveals New Lipid-Binding Proteins with a Putative Role in Lipid-Mediated Signaling.Front Plant Sci. 2016 Apr 28;7:563. doi: 10.3389/fpls.2016.00563. eCollection 2016. Front Plant Sci. 2016. PMID: 27200036 Free PMC article.
-
Drought and heat stress mediated activation of lipid signaling in plants: a critical review.Front Plant Sci. 2023 Aug 10;14:1216835. doi: 10.3389/fpls.2023.1216835. eCollection 2023. Front Plant Sci. 2023. PMID: 37636093 Free PMC article. Review.
-
Cytidinediphosphate diacylglycerol synthase-Mediated phosphatidic acid metabolism is crucial for early embryonic development of Arabidopsis.PLoS Genet. 2022 Jul 25;18(7):e1010320. doi: 10.1371/journal.pgen.1010320. eCollection 2022 Jul. PLoS Genet. 2022. PMID: 35877676 Free PMC article.
References
-
- Alvarez-Venegas R., Sadder M., Hlavacka A., Baluska F., Xia Y., Lu G., Firsov A., Sarath G., Moriyama H., Dubrovsky J.G., Avramova Z. (2006). The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. USA 103: 6049–6054 - PMC - PubMed
-
- Chen H., Nelson R.S., Sherwood J.L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16: 664–668, 670 - PubMed
-
- Di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., Morelli G. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10: 393–402 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

