Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds
- PMID: 24368787
- PMCID: PMC3903990
- DOI: 10.1105/tpc.113.119370
Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds
Abstract
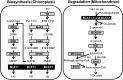
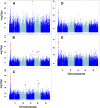
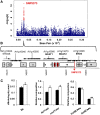
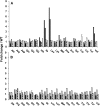
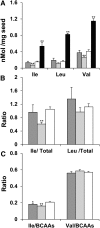
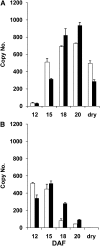
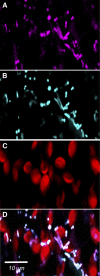
Branched-chain amino acids (BCAAs) are three of the nine essential amino acids in human and animal diets and are important for numerous processes in development and growth. However, seed BCAA levels in major crops are insufficient to meet dietary requirements, making genetic improvement for increased and balanced seed BCAAs an important nutritional target. Addressing this issue requires a better understanding of the genetics underlying seed BCAA content and composition. Here, a genome-wide association study and haplotype analysis for seed BCAA traits in Arabidopsis thaliana revealed a strong association with a chromosomal interval containing two branched-chain amino acid transferases, BCAT1 and BCAT2. Linkage analysis, reverse genetic approaches, and molecular complementation analysis demonstrated that allelic variation at BCAT2 is responsible for the natural variation of seed BCAAs in this interval. Complementation analysis of a bcat2 null mutant with two significantly different alleles from accessions Bayreuth-0 and Shahdara is consistent with BCAT2 contributing to natural variation in BCAA levels, glutamate recycling, and free amino acid homeostasis in seeds in an allele-dependent manner. The seed-specific phenotype of bcat2 null alleles, its strong transcription induction during late seed development, and its subcellular localization to the mitochondria are consistent with a unique, catabolic role for BCAT2 in BCAA metabolism in seeds.
Figures
References
-
- Angelovici R., Galili G., Fernie A.R., Fait A. (2010). Seed desiccation: A bridge between maturation and germination. Trends Plant Sci. 15: 211–218 - PubMed
-
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Larson T.R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I.A., Leaver C.J., Fernie A.R. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 - PMC - PubMed
-
- Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 - PubMed
-
- Barrett J.C., Fry B., Maller J., Daly M.J. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

