The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula
- PMID: 24368797
- PMCID: PMC3903991
- DOI: 10.1105/tpc.113.117788
The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula
Abstract
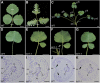
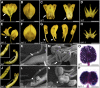
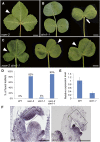
Leaf shape elaboration and organ separation are critical for plant morphogenesis. We characterized the developmental roles of lobed leaflet1 by analyzing a recessive mutant in the model legume Medicago truncatula. An ortholog of Arabidopsis thaliana argonaute7 (AGO7), Mt-AGO7/lobed leaflet1, is required for the biogenesis of a trans-acting short interfering RNA (ta-siRNA) to negatively regulate the expression of auxin response factors in M. truncatula. Loss of function in AGO7 results in pleiotropic phenotypes in different organs. The prominent phenotype of the ago7 mutant is lobed leaf margins and more widely spaced lateral organs, suggesting that the trans-acting siRNA3 (TAS3) pathway negatively regulates the formation of boundaries and the separation of lateral organs in M. truncatula. Genetic interaction analysis with the smooth leaf margin1 (slm1) mutant revealed that leaf margin formation is cooperatively regulated by the auxin/SLM1 (ortholog of Arabidopsis PIN-formed1) module, which influences the initiation of leaf margin teeth, and the TAS3 ta-siRNA pathway, which determines the degree of margin indentation. Further investigations showed that the TAS3 ta-siRNA pathway and no apical meristem (ortholog of Arabidopsis cup-shaped cotyledon) antagonistically regulate both leaf margin development and lateral organ separation, and the regulation is partially dependent on the auxin/SLM1 module.
Figures







Similar articles
-
Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development.Plant Cell. 2011 Jun;23(6):2106-24. doi: 10.1105/tpc.111.085464. Epub 2011 Jun 21. Plant Cell. 2011. PMID: 21693694 Free PMC article.
-
LATE MERISTEM IDENTITY1 regulates leaf margin development via the auxin transporter gene SMOOTH LEAF MARGIN1.Plant Physiol. 2021 Sep 4;187(1):218-235. doi: 10.1093/plphys/kiab268. Plant Physiol. 2021. PMID: 34618141 Free PMC article.
-
Auxin efflux transporter MtPIN10 regulates compound leaf and flower development in Medicago truncatula.Plant Signal Behav. 2011 Oct;6(10):1537-44. doi: 10.4161/psb.6.10.17326. Epub 2011 Oct 1. Plant Signal Behav. 2011. PMID: 21900740 Free PMC article.
-
Functional Genomics and Genetic Control of Compound Leaf Development in Medicago truncatula: An Overview.Methods Mol Biol. 2018;1822:197-203. doi: 10.1007/978-1-4939-8633-0_14. Methods Mol Biol. 2018. PMID: 30043306 Review.
-
Leaf Development in Medicago truncatula.Genes (Basel). 2022 Jul 5;13(7):1203. doi: 10.3390/genes13071203. Genes (Basel). 2022. PMID: 35885986 Free PMC article. Review.
Cited by
-
A class II KNOX gene, KNOX4, controls seed physical dormancy.Proc Natl Acad Sci U S A. 2016 Jun 21;113(25):6997-7002. doi: 10.1073/pnas.1601256113. Epub 2016 Jun 6. Proc Natl Acad Sci U S A. 2016. PMID: 27274062 Free PMC article.
-
Extensive Families of miRNAs and PHAS Loci in Norway Spruce Demonstrate the Origins of Complex phasiRNA Networks in Seed Plants.Mol Biol Evol. 2015 Nov;32(11):2905-18. doi: 10.1093/molbev/msv164. Epub 2015 Aug 28. Mol Biol Evol. 2015. PMID: 26318183 Free PMC article.
-
Genome-Wide Identification and Evolutionary Analysis of Argonaute Genes in Hexaploid Bread Wheat.Biomed Res Int. 2021 Jun 18;2021:9983858. doi: 10.1155/2021/9983858. eCollection 2021. Biomed Res Int. 2021. PMID: 34239939 Free PMC article.
-
Regulation of compound leaf development in mungbean (Vigna radiata L.) by CUP-SHAPED COTYLEDON/NO APICAL MERISTEM (CUC/NAM) gene.Planta. 2019 Mar;249(3):765-774. doi: 10.1007/s00425-018-3038-z. Epub 2018 Nov 2. Planta. 2019. PMID: 30390139
-
Ancient trans-Acting siRNAs Confer Robustness and Sensitivity onto the Auxin Response.Dev Cell. 2016 Feb 8;36(3):276-89. doi: 10.1016/j.devcel.2016.01.010. Dev Cell. 2016. PMID: 26859352 Free PMC article.
References
-
- Abdi H. (2007). Bonferroni and Sidak corrections for multiple comparisons. In Encyclopedia of Measurement and Statistics, N.J. Salkind, ed. (Thousand Oaks, California: Sage), pp. 103–107.
-
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932 - PubMed
-
- Aida M., Tasaka M. (2006). Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol. 9: 72–77 - PubMed
-
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40: 1136–1141 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

