CLASP2 interacts with p120-catenin and governs microtubule dynamics at adherens junctions
- PMID: 24368809
- PMCID: PMC3871427
- DOI: 10.1083/jcb.201306019
CLASP2 interacts with p120-catenin and governs microtubule dynamics at adherens junctions
Abstract
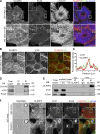
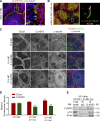
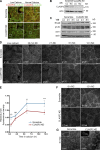
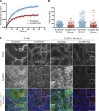
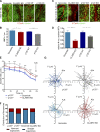
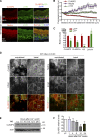
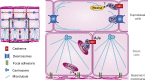
Classical cadherins and their connections with microtubules (MTs) are emerging as important determinants of cell adhesion. However, the functional relevance of such interactions and the molecular players that contribute to tissue architecture are still emerging. In this paper, we report that the MT plus end-binding protein CLASP2 localizes to adherens junctions (AJs) via direct interaction with p120-catenin (p120) in primary basal mouse keratinocytes. Reductions in the levels of p120 or CLASP2 decreased the localization of the other protein to cell-cell contacts and altered AJ dynamics and stability. These features were accompanied by decreased MT density and altered MT dynamics at intercellular junction sites. Interestingly, CLASP2 was enriched at the cortex of basal progenitor keratinocytes, in close localization to p120. Our findings suggest the existence of a new mechanism of MT targeting to AJs with potential functional implications in the maintenance of proper cell-cell adhesion in epidermal stem cells.
Figures


Comment in
-
Microtubules CLASP to Adherens Junctions in epidermal progenitor cells.Bioarchitecture. 2014 Jan-Feb;4(1):25-30. doi: 10.4161/bioa.28177. Epub 2014 Feb 12. Bioarchitecture. 2014. PMID: 24522006 Free PMC article.
Similar articles
-
Microtubules CLASP to Adherens Junctions in epidermal progenitor cells.Bioarchitecture. 2014 Jan-Feb;4(1):25-30. doi: 10.4161/bioa.28177. Epub 2014 Feb 12. Bioarchitecture. 2014. PMID: 24522006 Free PMC article.
-
Negative pressure induces p120-catenin-dependent adherens junction disassembly in keratinocytes during wound healing.Biochim Biophys Acta. 2016 Sep;1863(9):2212-20. doi: 10.1016/j.bbamcr.2016.05.017. Epub 2016 May 22. Biochim Biophys Acta. 2016. PMID: 27220534
-
Binding between the junctional proteins afadin and PLEKHA7 and implication in the formation of adherens junction in epithelial cells.J Biol Chem. 2013 Oct 11;288(41):29356-68. doi: 10.1074/jbc.M113.453464. Epub 2013 Aug 29. J Biol Chem. 2013. PMID: 23990464 Free PMC article.
-
Regulation of adherens junctions by Rho GTPases and p120-catenin.Arch Biochem Biophys. 2012 Aug 1;524(1):48-55. doi: 10.1016/j.abb.2012.04.019. Epub 2012 May 11. Arch Biochem Biophys. 2012. PMID: 22583808 Review.
-
Structure and biochemistry of cadherins and catenins.Cold Spring Harb Perspect Biol. 2009 Sep;1(3):a003053. doi: 10.1101/cshperspect.a003053. Cold Spring Harb Perspect Biol. 2009. PMID: 20066110 Free PMC article. Review.
Cited by
-
Involvement of CRF2 signaling in enterocyte differentiation.World J Gastroenterol. 2017 Jul 28;23(28):5127-5145. doi: 10.3748/wjg.v23.i28.5127. World J Gastroenterol. 2017. PMID: 28811708 Free PMC article.
-
Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin.Proc Natl Acad Sci U S A. 2014 Apr 22;111(16):5860-5. doi: 10.1073/pnas.1404133111. Epub 2014 Mar 26. Proc Natl Acad Sci U S A. 2014. PMID: 24706919 Free PMC article.
-
Microtubules CLASP to Adherens Junctions in epidermal progenitor cells.Bioarchitecture. 2014 Jan-Feb;4(1):25-30. doi: 10.4161/bioa.28177. Epub 2014 Feb 12. Bioarchitecture. 2014. PMID: 24522006 Free PMC article.
-
CLASP2 is involved in the EMT and early progression after transurethral resection of the bladder tumor.BMC Cancer. 2017 Feb 6;17(1):105. doi: 10.1186/s12885-017-3101-3. BMC Cancer. 2017. PMID: 28166762 Free PMC article.
-
Characterization of the CLASP2 Protein Interaction Network Identifies SOGA1 as a Microtubule-Associated Protein.Mol Cell Proteomics. 2017 Oct;16(10):1718-1735. doi: 10.1074/mcp.RA117.000011. Epub 2017 May 26. Mol Cell Proteomics. 2017. PMID: 28550165 Free PMC article.
References
-
- Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B.M., De Zeeuw C.I., Grosveld F., Galjart N. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 104:923–935 10.1016/S0092-8674(01)00288-4 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

