Calpain 2 activation of P-TEFb drives megakaryocyte morphogenesis and is disrupted by leukemogenic GATA1 mutation
- PMID: 24369834
- PMCID: PMC3892434
- DOI: 10.1016/j.devcel.2013.11.013
Calpain 2 activation of P-TEFb drives megakaryocyte morphogenesis and is disrupted by leukemogenic GATA1 mutation
Abstract
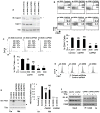
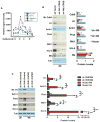
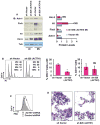
Megakaryocyte morphogenesis employs a "hypertrophy-like" developmental program that is dependent on P-TEFb kinase activation and cytoskeletal remodeling. P-TEFb activation classically occurs by a feedback-regulated process of signal-induced, reversible release of active Cdk9-cyclin T modules from large, inactive 7SK small nuclear ribonucleoprotein particle (snRNP) complexes. Here, we have identified an alternative pathway of irreversible P-TEFb activation in megakaryopoiesis that is mediated by dissolution of the 7SK snRNP complex. In this pathway, calpain 2 cleavage of the core 7SK snRNP component MePCE promoted P-TEFb release and consequent upregulation of a cohort of cytoskeleton remodeling factors, including α-actinin-1. In a subset of human megakaryocytic leukemias, the transcription factor GATA1 undergoes truncating mutation (GATA1s). Here, we linked the GATA1s mutation to defects in megakaryocytic upregulation of calpain 2 and of P-TEFb-dependent cytoskeletal remodeling factors. Restoring calpain 2 expression in GATA1s mutant megakaryocytes rescued normal development, implicating this morphogenetic pathway as a target in human leukemogenesis.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Megakaryocytic irreversible P-TEFb activation.Cell Cycle. 2014;13(12):1827-8. doi: 10.4161/cc.29324. Epub 2014 May 27. Cell Cycle. 2014. PMID: 24866976 Free PMC article. No abstract available.
References
-
- Bourquin JP, Subramanian A, Langebrake C, Reinhardt D, Bernard O, Ballerini P, Baruchel A, Cave H, Dastugue N, Hasle H, Kaspers GL, Lessard M, Michaux L, Vyas P, van Wering E, Zwaan CM, Golub TR, Orkin SH. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci USA. 2006;103:3339–3344. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

