Developing high-throughput HIV incidence assay with pyrosequencing platform
- PMID: 24371062
- PMCID: PMC3958066
- DOI: 10.1128/JVI.03128-13
Developing high-throughput HIV incidence assay with pyrosequencing platform
Abstract
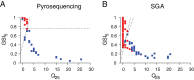
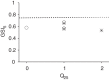
Human immunodeficiency virus (HIV) incidence is an important measure for monitoring the epidemic and evaluating the efficacy of intervention and prevention trials. This study developed a high-throughput, single-measure incidence assay by implementing a pyrosequencing platform. We devised a signal-masking bioinformatics pipeline, which yielded a process error rate of 5.8 × 10(-4) per base. The pipeline was then applied to analyze 18,434 envelope gene segments (HXB2 7212 to 7601) obtained from 12 incident and 24 chronic patients who had documented HIV-negative and/or -positive tests. The pyrosequencing data were cross-checked by using the single-genome-amplification (SGA) method to independently obtain 302 sequences from 13 patients. Using two genomic biomarkers that probe for the presence of similar sequences, the pyrosequencing platform correctly classified all 12 incident subjects (100% sensitivity) and 23 of 24 chronic subjects (96% specificity). One misclassified subject's chronic infection was correctly classified by conducting the same analysis with SGA data. The biomarkers were statistically associated across the two platforms, suggesting the assay's reproducibility and robustness. Sampling simulations showed that the biomarkers were tolerant of sequencing errors and template resampling, two factors most likely to affect the accuracy of pyrosequencing results. We observed comparable biomarker scores between AIDS and non-AIDS chronic patients (multivariate analysis of variance [MANOVA], P = 0.12), indicating that the stage of HIV disease itself does not affect the classification scheme. The high-throughput genomic HIV incidence marks a significant step toward determining incidence from a single measure in cross-sectional surveys.
Importance: Annual HIV incidence, the number of newly infected individuals within a year, is the key measure of monitoring the epidemic's rise and decline. Developing reliable assays differentiating recent from chronic infections has been a long-standing quest in the HIV community. Over the past 15 years, these assays have traditionally measured various HIV-specific antibodies, but recent technological advancements have expanded the diversity of proposed accurate, user-friendly, and financially viable tools. Here we designed a high-throughput genomic HIV incidence assay based on the signature imprinted in the HIV gene sequence population. By combining next-generation sequencing techniques with bioinformatics analysis, we demonstrated that genomic fingerprints are capable of distinguishing recently infected patients from chronically infected patients with high precision. Our high-throughput platform is expected to allow us to process many patients' samples from a single experiment, permitting the assay to be cost-effective for routine surveillance.
Figures

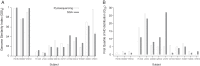
References
-
- Busch MP, Pilcher CD, Mastro TD, Kaldor J, Vercauteren G, Rodriguez W, Rousseau C, Rehle TM, Welte A, Averill MD, Garcia Calleja JM. 2010. Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS 24:2763–2771. 10.1097/QAD.0b013e32833f1142 - DOI - PubMed
-
- Brookmeyer R, Quinn TC. 1995. Estimation of current human immunodeficiency virus incidence rates from a cross-sectional survey using early diagnostic tests. Am. J. Epidemiol. 141:166–172 - PubMed
-
- Janssen RS, Satten GA, Stramer SL, Rawal BD, O'Brien TR, Weiblen BJ, Hecht FM, Jack N, Cleghorn FR, Kahn JO, Chesney MA, Busch MP. 1998. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 280:42–48. 10.1001/jama.280.1.42 - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

