NanoLuc reporter for dual luciferase imaging in living animals
- PMID: 24371848
- PMCID: PMC4144862
NanoLuc reporter for dual luciferase imaging in living animals
Abstract
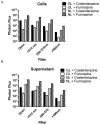
Bioluminescence imaging is widely used for cell-based assays and animal imaging studies in biomedical research and drug development, capitalizing on the high signal to background of this technique. A relatively small number of luciferases are available for imaging studies, substantially limiting the ability to image multiple molecular and cellular events, as done commonly with fluorescence imaging. To advance dual reporter bioluminescence molecular imaging, we tested a recently developed, adenosine triphosphate–independent luciferase enzyme from Oplophorus gracilirostris (NanoLuc [NL]) as a reporter for animal imaging. We demonstrated that NL could be imaged in superficial and deep tissues in living mice, although the detection of NL in deep tissues was limited by emission of predominantly blue light by this enzyme. Changes in bioluminescence from NL over time could be used to quantify tumor growth, and secreted NL was detectable in small volumes of serum. We combined NL and firefly luciferase reporters to quantify two key steps in transforming growth factor β signaling in intact cells and living mice, establishing a novel dual luciferase imaging strategy for quantifying signal transduction and drug targeting. Our results establish NL as a new reporter for bioluminescence imaging studies in intact cells and living mice that will expand imaging of signal transduction in normal physiology, disease, and drug development.
Figures
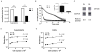
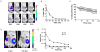
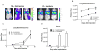

References
-
- Zhang L, Lee K, Bhojani M, et al. Molecular imaging of Akt kinase activity. Nat Med. 2007;13(9):1114–1119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
