Steps towards the synthetic biology of polyketide biosynthesis
- PMID: 24372666
- PMCID: PMC4237116
- DOI: 10.1111/1574-6968.12365
Steps towards the synthetic biology of polyketide biosynthesis
Abstract
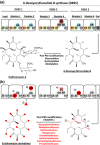
Nature is providing a bountiful pool of valuable secondary metabolites, many of which possess therapeutic properties. However, the discovery of new bioactive secondary metabolites is slowing down, at a time when the rise of multidrug-resistant pathogens and the realization of acute and long-term side effects of widely used drugs lead to an urgent need for new therapeutic agents. Approaches such as synthetic biology are promising to deliver a much-needed boost to secondary metabolite drug development through plug-and-play optimized hosts and refactoring novel or cryptic bacterial gene clusters. Here, we discuss this prospect focusing on one comprehensively studied class of clinically relevant bioactive molecules, the polyketides. Extensive efforts towards optimization and derivatization of compounds via combinatorial biosynthesis and classical engineering have elucidated the modularity, flexibility and promiscuity of polyketide biosynthetic enzymes. Hence, a synthetic biology approach can build upon a solid basis of guidelines and principles, while providing a new perspective towards the discovery and generation of novel and new-to-nature compounds. We discuss the lessons learned from the classical engineering of polyketide synthases and indicate their importance when attempting to engineer biosynthetic pathways using synthetic biology approaches for the introduction of novelty and overexpression of products in a controllable manner.
Keywords: combinatorial biosynthesis; drug discovery; plug-and-play biology; refactoring; secondary metabolites.
© 2013 The Authors FEMS Microbiology Letters published by John Wiley & Sons Ltd on behalf of Federation of European Microbiological Societies.
Figures

References
-
- Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Brana AF, Mendez C. Salas JA. Identification and expression of genes involved in biosynthesis of L-oleandrose and its intermediate L-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob Agents Chemother. 2000;44:1266–1275. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

